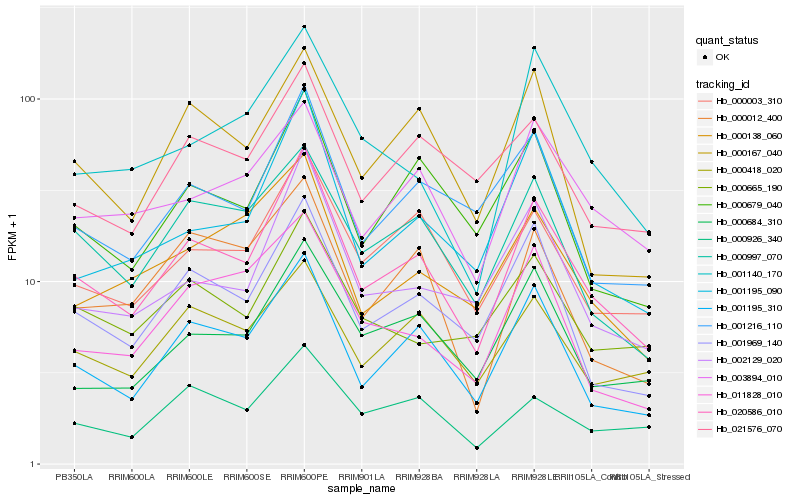
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_020586_010 |
0.0 |
- |
- |
Alpha-N-arabinofuranosidase 1 precursor, putative [Ricinus communis] |
| 2 |
Hb_001195_310 |
0.0848346235 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 3 |
Hb_001969_140 |
0.1017667955 |
- |
- |
PREDICTED: uncharacterized protein LOC105634929 [Jatropha curcas] |
| 4 |
Hb_002129_020 |
0.1102911094 |
- |
- |
PREDICTED: probable beta-1,4-xylosyltransferase IRX14 [Jatropha curcas] |
| 5 |
Hb_000003_310 |
0.1117613343 |
- |
- |
PREDICTED: glycosyltransferase-like KOBITO 1 [Jatropha curcas] |
| 6 |
Hb_001216_110 |
0.1137899535 |
- |
- |
PREDICTED: protein GPR107-like [Jatropha curcas] |
| 7 |
Hb_000679_040 |
0.1141172188 |
- |
- |
Cellulose synthase 1 [Theobroma cacao] |
| 8 |
Hb_000997_070 |
0.1190152997 |
- |
- |
PREDICTED: ureidoglycolate hydrolase isoform X1 [Jatropha curcas] |
| 9 |
Hb_001195_090 |
0.1191649112 |
- |
- |
PREDICTED: isoflavone reductase-like protein [Jatropha curcas] |
| 10 |
Hb_000684_310 |
0.1202475626 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g67720 [Jatropha curcas] |
| 11 |
Hb_003894_010 |
0.1259818609 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 12 |
Hb_021576_070 |
0.1288290305 |
- |
- |
hypothetical protein PRUPE_ppa002736mg [Prunus persica] |
| 13 |
Hb_000418_020 |
0.1297673457 |
- |
- |
PREDICTED: sulfhydryl oxidase 2 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000167_040 |
0.1301634892 |
- |
- |
PREDICTED: serine carboxypeptidase-like [Jatropha curcas] |
| 15 |
Hb_000926_340 |
0.1302255826 |
- |
- |
PREDICTED: EH domain-containing protein 1 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000665_190 |
0.1314540624 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000138_060 |
0.1319933929 |
- |
- |
PREDICTED: protein trichome birefringence [Jatropha curcas] |
| 18 |
Hb_001140_170 |
0.1320674505 |
- |
- |
PREDICTED: remorin [Jatropha curcas] |
| 19 |
Hb_000012_400 |
0.1321239606 |
- |
- |
PREDICTED: K(+) efflux antiporter 6 [Jatropha curcas] |
| 20 |
Hb_011828_010 |
0.1330438006 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g67720 [Jatropha curcas] |