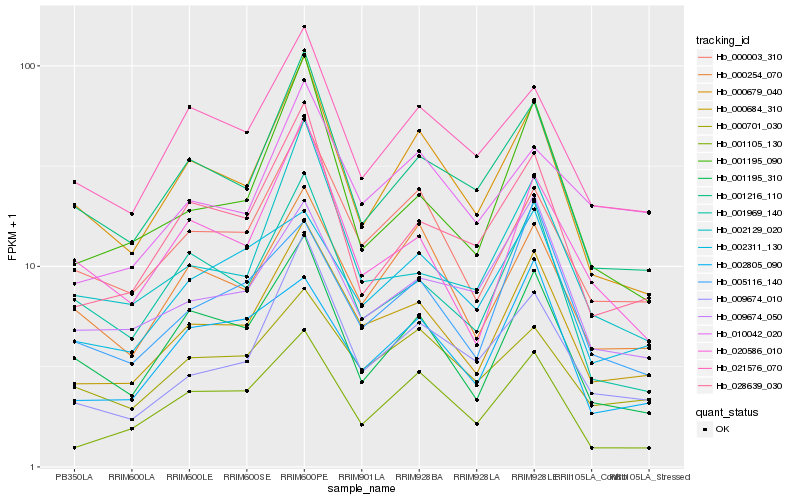
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000684_310 |
0.0 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g67720 [Jatropha curcas] |
| 2 |
Hb_000003_310 |
0.0928221941 |
- |
- |
PREDICTED: glycosyltransferase-like KOBITO 1 [Jatropha curcas] |
| 3 |
Hb_001216_110 |
0.1081057532 |
- |
- |
PREDICTED: protein GPR107-like [Jatropha curcas] |
| 4 |
Hb_009674_010 |
0.1081506408 |
- |
- |
AAEL007687-PA [Aedes aegypti] |
| 5 |
Hb_000679_040 |
0.10839585 |
- |
- |
Cellulose synthase 1 [Theobroma cacao] |
| 6 |
Hb_000254_070 |
0.1100327666 |
- |
- |
PREDICTED: protein disulfide isomerase-like 1-6 [Jatropha curcas] |
| 7 |
Hb_001105_130 |
0.1140959748 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase RPK2 [Jatropha curcas] |
| 8 |
Hb_028639_030 |
0.1142294247 |
- |
- |
PREDICTED: transmembrane 9 superfamily member 2-like [Jatropha curcas] |
| 9 |
Hb_001195_310 |
0.1149594383 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 10 |
Hb_021576_070 |
0.1151095909 |
- |
- |
hypothetical protein PRUPE_ppa002736mg [Prunus persica] |
| 11 |
Hb_005116_140 |
0.117042364 |
- |
- |
PREDICTED: RAN GTPase-activating protein 1 [Jatropha curcas] |
| 12 |
Hb_000701_030 |
0.1179082729 |
- |
- |
Sodium/hydrogen exchanger 6 -like protein [Gossypium arboreum] |
| 13 |
Hb_001195_090 |
0.1179617506 |
- |
- |
PREDICTED: isoflavone reductase-like protein [Jatropha curcas] |
| 14 |
Hb_009674_050 |
0.1195361419 |
- |
- |
Endosomal P24A protein precursor, putative [Ricinus communis] |
| 15 |
Hb_002129_020 |
0.1196313317 |
- |
- |
PREDICTED: probable beta-1,4-xylosyltransferase IRX14 [Jatropha curcas] |
| 16 |
Hb_020586_010 |
0.1202475626 |
- |
- |
Alpha-N-arabinofuranosidase 1 precursor, putative [Ricinus communis] |
| 17 |
Hb_001969_140 |
0.1218848338 |
- |
- |
PREDICTED: uncharacterized protein LOC105634929 [Jatropha curcas] |
| 18 |
Hb_002311_130 |
0.1225029377 |
- |
- |
PREDICTED: importin subunit alpha-2 isoform X2 [Jatropha curcas] |
| 19 |
Hb_002805_090 |
0.1254697232 |
- |
- |
Ran GTPase binding protein, putative [Ricinus communis] |
| 20 |
Hb_010042_020 |
0.1272035772 |
- |
- |
UDP-n-acteylglucosamine pyrophosphorylase, putative [Ricinus communis] |