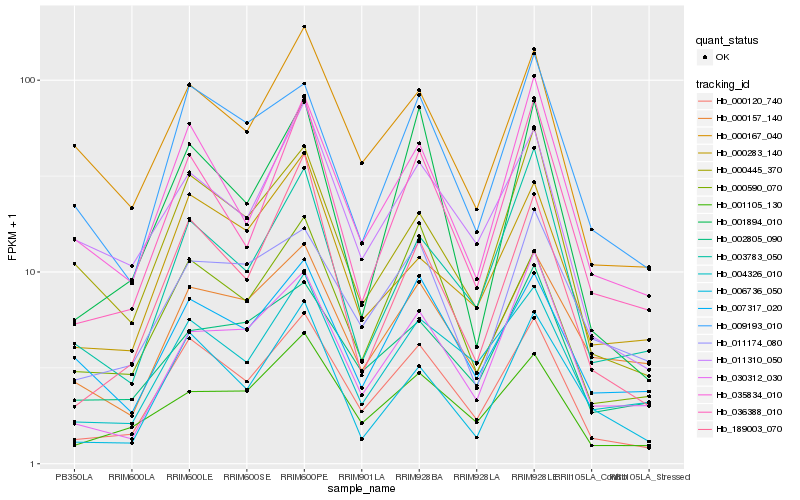
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000120_740 |
0.0 |
desease resistance |
Gene Name: VARLMGL |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_036388_010 |
0.109430247 |
- |
- |
PREDICTED: formate--tetrahydrofolate ligase [Populus euphratica] |
| 3 |
Hb_004326_010 |
0.1099495019 |
- |
- |
cationic amino acid transporter, putative [Ricinus communis] |
| 4 |
Hb_001105_130 |
0.1119858248 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase RPK2 [Jatropha curcas] |
| 5 |
Hb_035834_010 |
0.1120235556 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 6 |
Hb_000590_070 |
0.1142129561 |
- |
- |
PREDICTED: beta-hexosaminidase 1 [Jatropha curcas] |
| 7 |
Hb_009193_010 |
0.116378997 |
- |
- |
PREDICTED: PLASMODESMATA CALLOSE-BINDING PROTEIN 3 [Jatropha curcas] |
| 8 |
Hb_011310_050 |
0.1222655974 |
- |
- |
PREDICTED: cellulose synthase A catalytic subunit 1 [UDP-forming] [Jatropha curcas] |
| 9 |
Hb_000445_370 |
0.1225536457 |
- |
- |
PREDICTED: probable methyltransferase PMT24 [Jatropha curcas] |
| 10 |
Hb_189003_070 |
0.1228949239 |
- |
- |
PREDICTED: peptide-N4-(N-acetyl-beta-glucosaminyl)asparagine amidase A-like [Jatropha curcas] |
| 11 |
Hb_000167_040 |
0.1240434461 |
- |
- |
PREDICTED: serine carboxypeptidase-like [Jatropha curcas] |
| 12 |
Hb_000283_140 |
0.1258050553 |
- |
- |
PREDICTED: transcriptional corepressor LEUNIG-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_006736_050 |
0.127702108 |
- |
- |
hypothetical protein POPTR_0006s04450g [Populus trichocarpa] |
| 14 |
Hb_001894_010 |
0.1295664616 |
- |
- |
PREDICTED: probable polygalacturonase non-catalytic subunit JP650 [Jatropha curcas] |
| 15 |
Hb_030312_030 |
0.1301836783 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP40 isoform X1 [Jatropha curcas] |
| 16 |
Hb_007317_020 |
0.1310466276 |
- |
- |
PREDICTED: transmembrane 9 superfamily member 5 isoform X1 [Jatropha curcas] |
| 17 |
Hb_011174_080 |
0.1315642144 |
- |
- |
PREDICTED: sulfite oxidase [Jatropha curcas] |
| 18 |
Hb_003783_050 |
0.1316258454 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_002805_090 |
0.1333142229 |
- |
- |
Ran GTPase binding protein, putative [Ricinus communis] |
| 20 |
Hb_000157_140 |
0.1338713229 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |