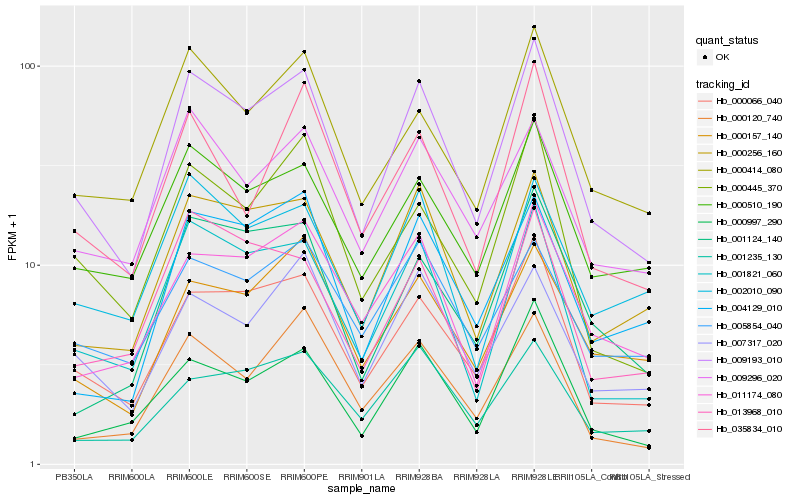
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009193_010 |
0.0 |
- |
- |
PREDICTED: PLASMODESMATA CALLOSE-BINDING PROTEIN 3 [Jatropha curcas] |
| 2 |
Hb_000066_040 |
0.0762024919 |
- |
- |
PREDICTED: CDPK-related kinase 7-like [Populus euphratica] |
| 3 |
Hb_001821_060 |
0.0841647749 |
- |
- |
PREDICTED: C-type lectin receptor-like tyrosine-protein kinase At1g52310 [Jatropha curcas] |
| 4 |
Hb_000256_160 |
0.0958640705 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox protein knotted-1-like 3-like isoform X1 [Glycine max] |
| 5 |
Hb_000510_190 |
0.1020955484 |
- |
- |
glutathione reductase [Hevea brasiliensis] |
| 6 |
Hb_000414_080 |
0.1024085912 |
- |
- |
PREDICTED: triosephosphate isomerase, chloroplastic [Jatropha curcas] |
| 7 |
Hb_000997_290 |
0.1083865168 |
- |
- |
PREDICTED: acylamino-acid-releasing enzyme [Jatropha curcas] |
| 8 |
Hb_000445_370 |
0.1087972842 |
- |
- |
PREDICTED: probable methyltransferase PMT24 [Jatropha curcas] |
| 9 |
Hb_009296_020 |
0.1090005298 |
desease resistance |
Gene Name: ATP-synt_ab_N |
PREDICTED: ATP synthase subunit beta, mitochondrial-like [Jatropha curcas] |
| 10 |
Hb_007317_020 |
0.1112918532 |
- |
- |
PREDICTED: transmembrane 9 superfamily member 5 isoform X1 [Jatropha curcas] |
| 11 |
Hb_011174_080 |
0.1119360834 |
- |
- |
PREDICTED: sulfite oxidase [Jatropha curcas] |
| 12 |
Hb_000157_140 |
0.113845246 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 13 |
Hb_000120_740 |
0.116378997 |
desease resistance |
Gene Name: VARLMGL |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_004129_010 |
0.1176736249 |
- |
- |
aspartate aminotransferase, putative [Ricinus communis] |
| 15 |
Hb_001124_140 |
0.1204084265 |
- |
- |
PREDICTED: inactive beta-amylase 9 [Jatropha curcas] |
| 16 |
Hb_002010_090 |
0.1205939515 |
- |
- |
PREDICTED: aminomethyltransferase, mitochondrial [Jatropha curcas] |
| 17 |
Hb_005854_040 |
0.1210160998 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_001235_130 |
0.1227873875 |
- |
- |
- |
| 19 |
Hb_035834_010 |
0.1231868269 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 20 |
Hb_013968_010 |
0.1233023497 |
- |
- |
PREDICTED: U4/U6 small nuclear ribonucleoprotein Prp3 [Jatropha curcas] |