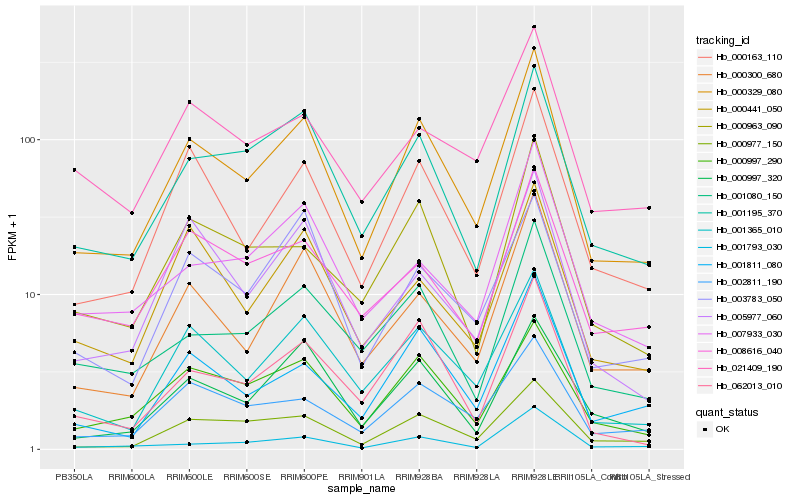
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000329_080 |
0.0 |
- |
- |
PREDICTED: transketolase, chloroplastic [Jatropha curcas] |
| 2 |
Hb_001365_010 |
0.0939947434 |
- |
- |
hypothetical protein JCGZ_02762 [Jatropha curcas] |
| 3 |
Hb_000163_110 |
0.094427362 |
- |
- |
PREDICTED: blue-light photoreceptor PHR2 [Jatropha curcas] |
| 4 |
Hb_001195_370 |
0.1050088576 |
- |
- |
- |
| 5 |
Hb_000300_680 |
0.1062793428 |
- |
- |
PREDICTED: uncharacterized protein LOC105632625 isoform X1 [Jatropha curcas] |
| 6 |
Hb_001811_080 |
0.1070360531 |
- |
- |
PREDICTED: fructose-1,6-bisphosphatase, cytosolic [Jatropha curcas] |
| 7 |
Hb_000977_150 |
0.1102107175 |
- |
- |
latex cyanogenic beta glucosidase [Hevea brasiliensis] |
| 8 |
Hb_002811_190 |
0.1111500799 |
- |
- |
tRNA pseudouridine synthase d, putative [Ricinus communis] |
| 9 |
Hb_000997_290 |
0.1113000045 |
- |
- |
PREDICTED: acylamino-acid-releasing enzyme [Jatropha curcas] |
| 10 |
Hb_062013_010 |
0.1182965354 |
- |
- |
phosphoprotein phosphatase, putative [Ricinus communis] |
| 11 |
Hb_001080_150 |
0.1188256589 |
- |
- |
PREDICTED: Niemann-Pick C1 protein-like [Jatropha curcas] |
| 12 |
Hb_000997_320 |
0.1200205479 |
- |
- |
PREDICTED: acylamino-acid-releasing enzyme-like [Malus domestica] |
| 13 |
Hb_000963_090 |
0.1205985646 |
rubber biosynthesis |
Gene Name: 4-hydroxy-3-methylbut-2-enyl diphosphate reductase |
4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase [Hevea brasiliensis] |
| 14 |
Hb_001793_030 |
0.1353497446 |
- |
- |
Cysteine-rich RLK (RECEPTOR-like protein kinase) 8 [Theobroma cacao] |
| 15 |
Hb_005977_060 |
0.1368631398 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_021409_190 |
0.1371979451 |
- |
- |
PREDICTED: luminal-binding protein 5-like [Jatropha curcas] |
| 17 |
Hb_007933_030 |
0.1374828422 |
- |
- |
PREDICTED: uncharacterized protein LOC105639932 [Jatropha curcas] |
| 18 |
Hb_000441_050 |
0.1375615117 |
- |
- |
PREDICTED: 187-kDa microtubule-associated protein AIR9 [Jatropha curcas] |
| 19 |
Hb_003783_050 |
0.1385393346 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_008616_040 |
0.1385518907 |
- |
- |
PREDICTED: RNA-binding protein CP33, chloroplastic [Jatropha curcas] |