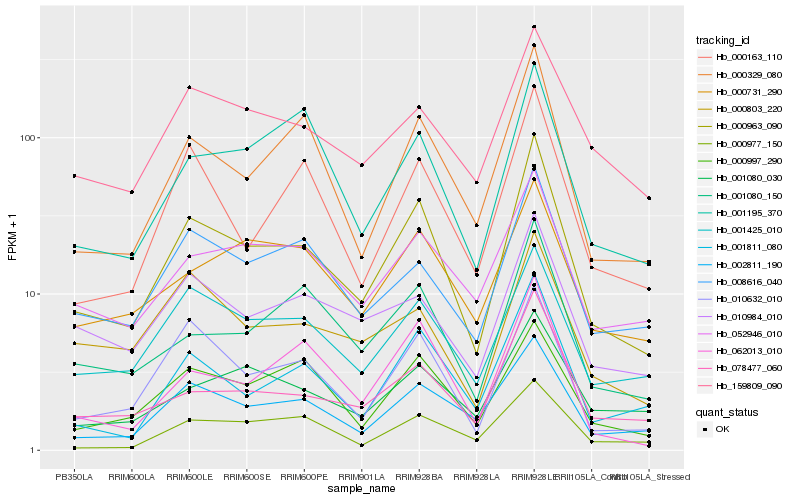
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000963_090 |
0.0 |
rubber biosynthesis |
Gene Name: 4-hydroxy-3-methylbut-2-enyl diphosphate reductase |
4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase [Hevea brasiliensis] |
| 2 |
Hb_010632_010 |
0.1159415535 |
- |
- |
PREDICTED: bifunctional dethiobiotin synthetase/7,8-diamino-pelargonic acid aminotransferase, mitochondrial [Jatropha curcas] |
| 3 |
Hb_078477_060 |
0.119299223 |
- |
- |
PREDICTED: histone-lysine N-methyltransferase ATX4 [Jatropha curcas] |
| 4 |
Hb_000329_080 |
0.1205985646 |
- |
- |
PREDICTED: transketolase, chloroplastic [Jatropha curcas] |
| 5 |
Hb_002811_190 |
0.1213713145 |
- |
- |
tRNA pseudouridine synthase d, putative [Ricinus communis] |
| 6 |
Hb_001811_080 |
0.1229178795 |
- |
- |
PREDICTED: fructose-1,6-bisphosphatase, cytosolic [Jatropha curcas] |
| 7 |
Hb_001080_150 |
0.1303726458 |
- |
- |
PREDICTED: Niemann-Pick C1 protein-like [Jatropha curcas] |
| 8 |
Hb_001425_010 |
0.1309967294 |
- |
- |
sugar transporter, putative [Ricinus communis] |
| 9 |
Hb_008616_040 |
0.1358498118 |
- |
- |
PREDICTED: RNA-binding protein CP33, chloroplastic [Jatropha curcas] |
| 10 |
Hb_062013_010 |
0.1363226762 |
- |
- |
phosphoprotein phosphatase, putative [Ricinus communis] |
| 11 |
Hb_000163_110 |
0.1402482667 |
- |
- |
PREDICTED: blue-light photoreceptor PHR2 [Jatropha curcas] |
| 12 |
Hb_000997_290 |
0.1410172521 |
- |
- |
PREDICTED: acylamino-acid-releasing enzyme [Jatropha curcas] |
| 13 |
Hb_052946_010 |
0.1437782824 |
- |
- |
PREDICTED: uncharacterized protein LOC105639103 [Jatropha curcas] |
| 14 |
Hb_010984_010 |
0.1444585596 |
- |
- |
PREDICTED: translation factor GUF1 homolog, chloroplastic isoform X1 [Jatropha curcas] |
| 15 |
Hb_159809_090 |
0.1485369409 |
rubber biosynthesis |
Gene Name: 4-hydroxy-3-methylbut-2-enyl diphosphate reductase |
4-hydroxy-3-methylbut-2-enyl diphosphate reductase [Hevea brasiliensis] |
| 16 |
Hb_001195_370 |
0.1493235624 |
- |
- |
- |
| 17 |
Hb_000977_150 |
0.1498401622 |
- |
- |
latex cyanogenic beta glucosidase [Hevea brasiliensis] |
| 18 |
Hb_000803_220 |
0.1511049848 |
- |
- |
PREDICTED: magnesium-chelatase subunit ChlD, chloroplastic [Jatropha curcas] |
| 19 |
Hb_001080_030 |
0.151369326 |
- |
- |
AMP dependent ligase, putative [Ricinus communis] |
| 20 |
Hb_000731_290 |
0.15278061 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |