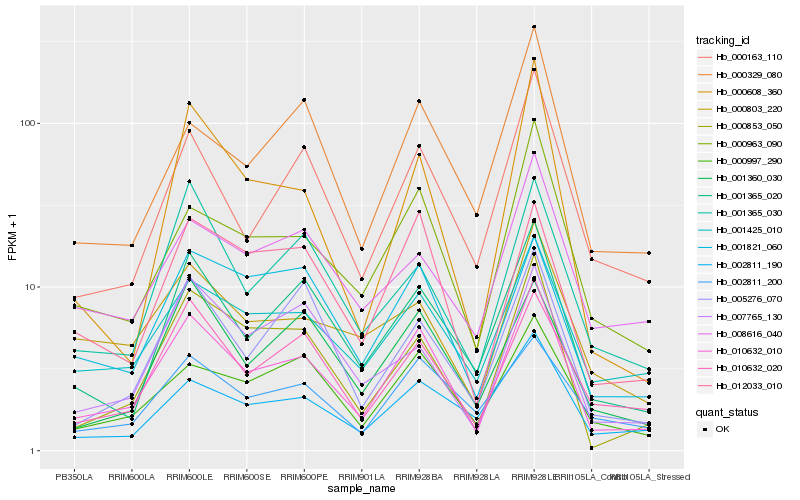
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010632_010 |
0.0 |
- |
- |
PREDICTED: bifunctional dethiobiotin synthetase/7,8-diamino-pelargonic acid aminotransferase, mitochondrial [Jatropha curcas] |
| 2 |
Hb_000963_090 |
0.1159415535 |
rubber biosynthesis |
Gene Name: 4-hydroxy-3-methylbut-2-enyl diphosphate reductase |
4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase [Hevea brasiliensis] |
| 3 |
Hb_001365_020 |
0.1230433247 |
- |
- |
hypothetical protein PRUPE_ppa000927mg [Prunus persica] |
| 4 |
Hb_000163_110 |
0.123902379 |
- |
- |
PREDICTED: blue-light photoreceptor PHR2 [Jatropha curcas] |
| 5 |
Hb_000853_050 |
0.125527147 |
- |
- |
PREDICTED: putative GTP diphosphokinase RSH1, chloroplastic isoform X1 [Jatropha curcas] |
| 6 |
Hb_001425_010 |
0.1265571036 |
- |
- |
sugar transporter, putative [Ricinus communis] |
| 7 |
Hb_010632_020 |
0.1271490958 |
- |
- |
hypothetical protein POPTR_0006s185902g, partial [Populus trichocarpa] |
| 8 |
Hb_000997_290 |
0.1324708661 |
- |
- |
PREDICTED: acylamino-acid-releasing enzyme [Jatropha curcas] |
| 9 |
Hb_002811_190 |
0.1364223696 |
- |
- |
tRNA pseudouridine synthase d, putative [Ricinus communis] |
| 10 |
Hb_012033_010 |
0.1372063576 |
- |
- |
PREDICTED: histone H1.2-like [Jatropha curcas] |
| 11 |
Hb_001360_030 |
0.1436436416 |
- |
- |
PREDICTED: ACT domain-containing protein ACR10 [Jatropha curcas] |
| 12 |
Hb_005276_070 |
0.1460525456 |
- |
- |
PREDICTED: RAN GTPase-activating protein 1 [Jatropha curcas] |
| 13 |
Hb_000329_080 |
0.1484021794 |
- |
- |
PREDICTED: transketolase, chloroplastic [Jatropha curcas] |
| 14 |
Hb_001821_060 |
0.1488078563 |
- |
- |
PREDICTED: C-type lectin receptor-like tyrosine-protein kinase At1g52310 [Jatropha curcas] |
| 15 |
Hb_001365_030 |
0.1490831685 |
- |
- |
PREDICTED: uncharacterized protein LOC102609547 [Citrus sinensis] |
| 16 |
Hb_002811_200 |
0.1500380836 |
- |
- |
PREDICTED: protein IQ-DOMAIN 14-like [Jatropha curcas] |
| 17 |
Hb_000803_220 |
0.1520983553 |
- |
- |
PREDICTED: magnesium-chelatase subunit ChlD, chloroplastic [Jatropha curcas] |
| 18 |
Hb_000608_360 |
0.15269677 |
- |
- |
calreticulin, putative [Ricinus communis] |
| 19 |
Hb_007765_130 |
0.1555713867 |
- |
- |
PREDICTED: UPF0503 protein At3g09070, chloroplastic [Jatropha curcas] |
| 20 |
Hb_008616_040 |
0.1556447982 |
- |
- |
PREDICTED: RNA-binding protein CP33, chloroplastic [Jatropha curcas] |