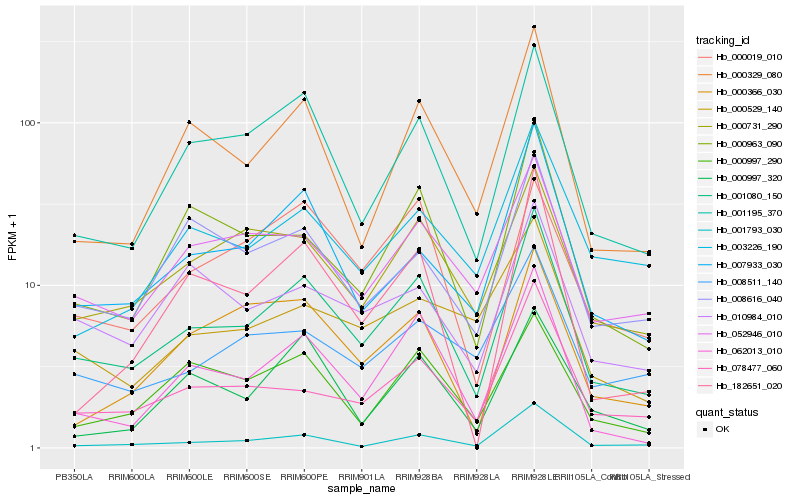
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001080_150 |
0.0 |
- |
- |
PREDICTED: Niemann-Pick C1 protein-like [Jatropha curcas] |
| 2 |
Hb_001195_370 |
0.1068263166 |
- |
- |
- |
| 3 |
Hb_062013_010 |
0.1188127602 |
- |
- |
phosphoprotein phosphatase, putative [Ricinus communis] |
| 4 |
Hb_000329_080 |
0.1188256589 |
- |
- |
PREDICTED: transketolase, chloroplastic [Jatropha curcas] |
| 5 |
Hb_007933_030 |
0.1237741318 |
- |
- |
PREDICTED: uncharacterized protein LOC105639932 [Jatropha curcas] |
| 6 |
Hb_000963_090 |
0.1303726458 |
rubber biosynthesis |
Gene Name: 4-hydroxy-3-methylbut-2-enyl diphosphate reductase |
4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase [Hevea brasiliensis] |
| 7 |
Hb_078477_060 |
0.1329262913 |
- |
- |
PREDICTED: histone-lysine N-methyltransferase ATX4 [Jatropha curcas] |
| 8 |
Hb_001793_030 |
0.1338671076 |
- |
- |
Cysteine-rich RLK (RECEPTOR-like protein kinase) 8 [Theobroma cacao] |
| 9 |
Hb_000731_290 |
0.1354322475 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 10 |
Hb_052946_010 |
0.1363417672 |
- |
- |
PREDICTED: uncharacterized protein LOC105639103 [Jatropha curcas] |
| 11 |
Hb_000997_290 |
0.1472502565 |
- |
- |
PREDICTED: acylamino-acid-releasing enzyme [Jatropha curcas] |
| 12 |
Hb_008616_040 |
0.1497310577 |
- |
- |
PREDICTED: RNA-binding protein CP33, chloroplastic [Jatropha curcas] |
| 13 |
Hb_000019_010 |
0.1511175165 |
- |
- |
PREDICTED: long chain base biosynthesis protein 2a [Jatropha curcas] |
| 14 |
Hb_010984_010 |
0.1519927823 |
- |
- |
PREDICTED: translation factor GUF1 homolog, chloroplastic isoform X1 [Jatropha curcas] |
| 15 |
Hb_182651_020 |
0.1531085885 |
- |
- |
alcohol dehydrogenase, putative [Ricinus communis] |
| 16 |
Hb_008511_140 |
0.1533247312 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105632314 [Jatropha curcas] |
| 17 |
Hb_003226_190 |
0.1550230367 |
- |
- |
PREDICTED: blue-light photoreceptor PHR2 [Jatropha curcas] |
| 18 |
Hb_000529_140 |
0.1554530623 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000366_030 |
0.1555545393 |
- |
- |
PREDICTED: 2-alkenal reductase (NADP(+)-dependent)-like [Jatropha curcas] |
| 20 |
Hb_000997_320 |
0.1558295206 |
- |
- |
PREDICTED: acylamino-acid-releasing enzyme-like [Malus domestica] |