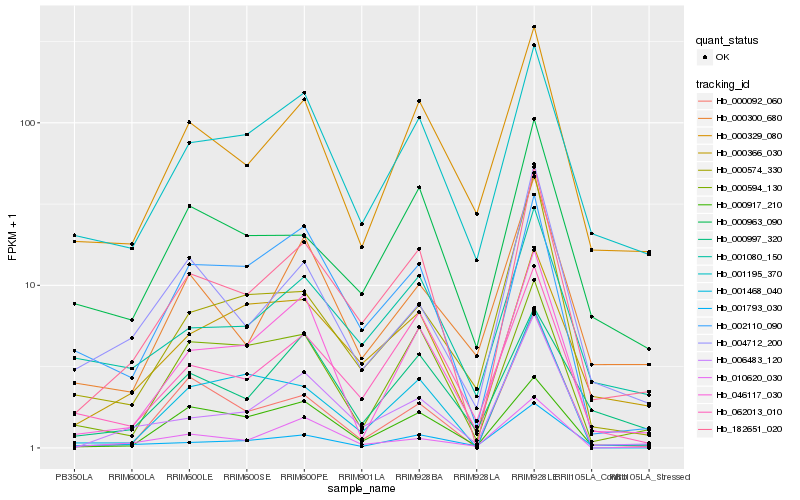
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_182651_020 |
0.0 |
- |
- |
alcohol dehydrogenase, putative [Ricinus communis] |
| 2 |
Hb_006483_120 |
0.1368121497 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 3 |
Hb_062013_010 |
0.1461192762 |
- |
- |
phosphoprotein phosphatase, putative [Ricinus communis] |
| 4 |
Hb_000366_030 |
0.1492114893 |
- |
- |
PREDICTED: 2-alkenal reductase (NADP(+)-dependent)-like [Jatropha curcas] |
| 5 |
Hb_001080_150 |
0.1531085885 |
- |
- |
PREDICTED: Niemann-Pick C1 protein-like [Jatropha curcas] |
| 6 |
Hb_000329_080 |
0.1543200298 |
- |
- |
PREDICTED: transketolase, chloroplastic [Jatropha curcas] |
| 7 |
Hb_001195_370 |
0.1572603759 |
- |
- |
- |
| 8 |
Hb_046117_030 |
0.1603522934 |
- |
- |
sialyltransferase, putative [Ricinus communis] |
| 9 |
Hb_000963_090 |
0.1616454568 |
rubber biosynthesis |
Gene Name: 4-hydroxy-3-methylbut-2-enyl diphosphate reductase |
4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase [Hevea brasiliensis] |
| 10 |
Hb_000917_210 |
0.1618310553 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000594_130 |
0.1629917529 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 12 |
Hb_002110_090 |
0.166070107 |
- |
- |
PREDICTED: transport inhibitor response 1-like protein [Jatropha curcas] |
| 13 |
Hb_004712_200 |
0.1696058485 |
- |
- |
hypothetical protein CICLE_v10010292mg [Citrus clementina] |
| 14 |
Hb_001468_040 |
0.1719260554 |
- |
- |
Uncharacterized protein TCM_024268 [Theobroma cacao] |
| 15 |
Hb_000997_320 |
0.1738111784 |
- |
- |
PREDICTED: acylamino-acid-releasing enzyme-like [Malus domestica] |
| 16 |
Hb_001793_030 |
0.1760519366 |
- |
- |
Cysteine-rich RLK (RECEPTOR-like protein kinase) 8 [Theobroma cacao] |
| 17 |
Hb_000300_680 |
0.1762223637 |
- |
- |
PREDICTED: uncharacterized protein LOC105632625 isoform X1 [Jatropha curcas] |
| 18 |
Hb_010620_030 |
0.1791409216 |
- |
- |
beta-amyrin synthase [Euphorbia tirucalli] |
| 19 |
Hb_000092_060 |
0.180535428 |
- |
- |
PREDICTED: uncharacterized protein LOC105646505 [Jatropha curcas] |
| 20 |
Hb_000574_330 |
0.1818865765 |
- |
- |
PREDICTED: clustered mitochondria protein homolog [Jatropha curcas] |