| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
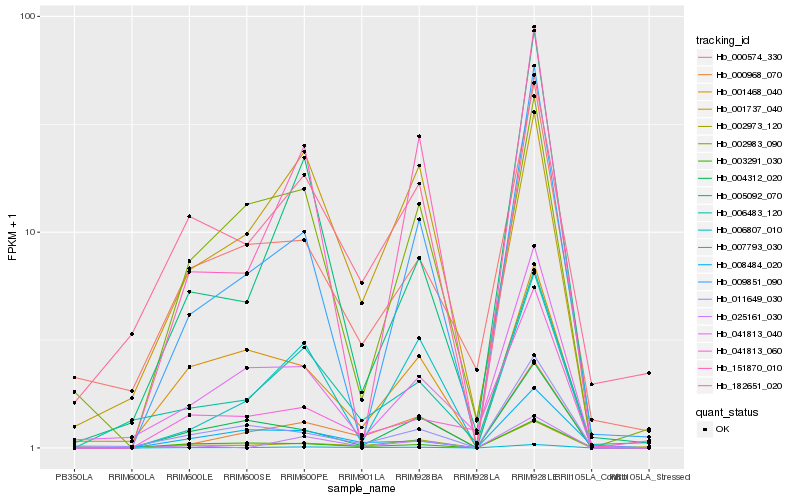
Hb_007793_030 |
0.0 |
- |
- |
OSJNBa0074B10.6 [Oryza sativa Japonica Group] |
| 2 |
Hb_000968_070 |
0.1229754039 |
- |
- |
- |
| 3 |
Hb_006483_120 |
0.1693339549 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 4 |
Hb_008484_020 |
0.1790907534 |
- |
- |
PREDICTED: uncharacterized protein LOC104106381 [Nicotiana tomentosiformis] |
| 5 |
Hb_003291_030 |
0.1834395038 |
- |
- |
Gag-pol protein, putative [Solanum demissum] |
| 6 |
Hb_182651_020 |
0.1849426145 |
- |
- |
alcohol dehydrogenase, putative [Ricinus communis] |
| 7 |
Hb_002973_120 |
0.1862855223 |
- |
- |
Uncharacterized protein TCM_024268 [Theobroma cacao] |
| 8 |
Hb_005092_070 |
0.1996533306 |
- |
- |
Oligopeptide transporter, putative [Ricinus communis] |
| 9 |
Hb_000574_330 |
0.2006165712 |
- |
- |
PREDICTED: clustered mitochondria protein homolog [Jatropha curcas] |
| 10 |
Hb_004312_020 |
0.201905109 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_011649_030 |
0.2047313488 |
- |
- |
hypothetical protein [Vitis hybrid cultivar] |
| 12 |
Hb_025161_030 |
0.2047557424 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 isoform X1 [Jatropha curcas] |
| 13 |
Hb_001468_040 |
0.2051145197 |
- |
- |
Uncharacterized protein TCM_024268 [Theobroma cacao] |
| 14 |
Hb_041813_060 |
0.2072392656 |
- |
- |
ribosomal protein L10 (mitochondrion) [Ricinus communis] |
| 15 |
Hb_009851_090 |
0.2080509848 |
- |
- |
Protein ABIL2, putative [Ricinus communis] |
| 16 |
Hb_006807_010 |
0.208261974 |
- |
- |
hypothetical protein JCGZ_20881 [Jatropha curcas] |
| 17 |
Hb_001737_040 |
0.2098804539 |
- |
- |
PREDICTED: uncharacterized protein LOC104100193 [Nicotiana tomentosiformis] |
| 18 |
Hb_041813_040 |
0.2105666912 |
- |
- |
NADH2 dehydrogenase (ubiquinone) (EC 1.6.5.3) chain 5, truncated - sugar beet mitochondrion (mitochondrion) [Beta vulgaris subsp. vulgaris] |
| 19 |
Hb_151870_010 |
0.211018743 |
- |
- |
Metalloendoproteinase 1 precursor, putative [Ricinus communis] |
| 20 |
Hb_002983_090 |
0.2116790848 |
- |
- |
Altered inheritance of mitochondria protein 32 [Glycine soja] |