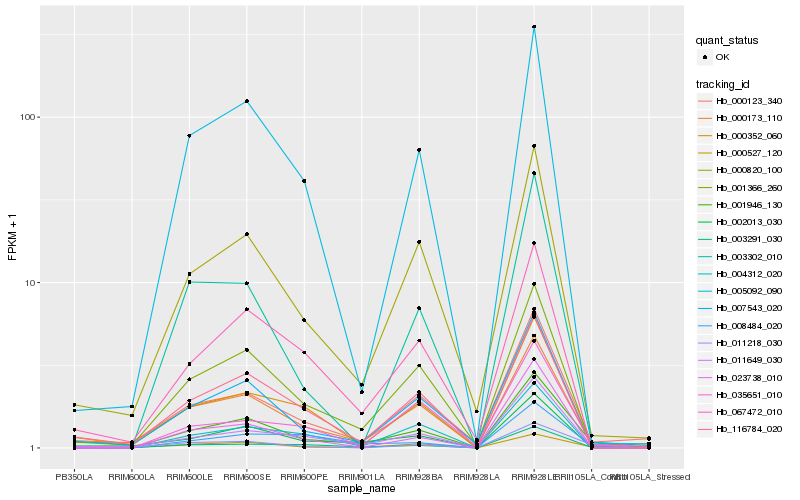
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001946_130 |
0.0 |
- |
- |
- |
| 2 |
Hb_003291_030 |
0.1351622681 |
- |
- |
Gag-pol protein, putative [Solanum demissum] |
| 3 |
Hb_001366_260 |
0.1386580455 |
- |
- |
T4.15 [Malus x robusta] |
| 4 |
Hb_007543_020 |
0.1503093414 |
- |
- |
PREDICTED: uncharacterized protein LOC104229533, partial [Nicotiana sylvestris] |
| 5 |
Hb_011649_030 |
0.1504505373 |
- |
- |
hypothetical protein [Vitis hybrid cultivar] |
| 6 |
Hb_000820_100 |
0.1525326685 |
- |
- |
RNA-directed DNA polymerase [Arachis hypogaea] |
| 7 |
Hb_000527_120 |
0.1531373141 |
- |
- |
hypothetical protein VITISV_027174 [Vitis vinifera] |
| 8 |
Hb_005092_090 |
0.1562761003 |
- |
- |
cold regulated protein [Manihot esculenta] |
| 9 |
Hb_000123_340 |
0.1601811265 |
- |
- |
PREDICTED: uncharacterized protein LOC105789586 [Gossypium raimondii] |
| 10 |
Hb_000173_110 |
0.1607440182 |
- |
- |
putative RNA-directed DNA polymerase (Reverse transcriptase) [Malus domestica] |
| 11 |
Hb_008484_020 |
0.1623852488 |
- |
- |
PREDICTED: uncharacterized protein LOC104106381 [Nicotiana tomentosiformis] |
| 12 |
Hb_004312_020 |
0.1625389244 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_011218_030 |
0.1654747447 |
- |
- |
hypothetical protein VITISV_002891 [Vitis vinifera] |
| 14 |
Hb_000352_060 |
0.166137284 |
- |
- |
RNA-directed DNA polymerase [Arachis hypogaea] |
| 15 |
Hb_003302_010 |
0.1663466017 |
- |
- |
PREDICTED: alpha-dioxygenase 1 [Jatropha curcas] |
| 16 |
Hb_023738_010 |
0.1722172774 |
- |
- |
Concanavalin A-like lectin protein kinase family protein [Theobroma cacao] |
| 17 |
Hb_067472_010 |
0.1743161218 |
- |
- |
Uncharacterized protein TCM_024268 [Theobroma cacao] |
| 18 |
Hb_035651_010 |
0.1743920959 |
- |
- |
receptor-kinase, putative [Ricinus communis] |
| 19 |
Hb_116784_020 |
0.1750220113 |
- |
- |
PREDICTED: uncharacterized protein LOC104898613 [Beta vulgaris subsp. vulgaris] |
| 20 |
Hb_002013_030 |
0.1761214236 |
- |
- |
PREDICTED: uncharacterized protein LOC104799692 [Tarenaya hassleriana] |