| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
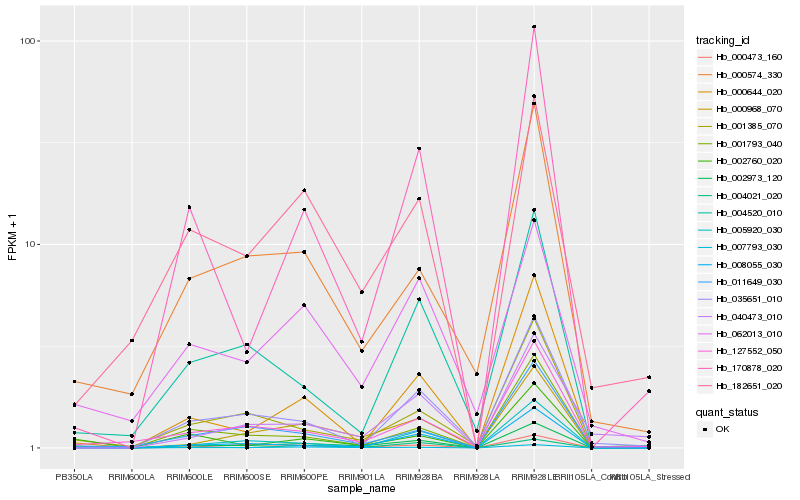
Hb_002973_120 |
0.0 |
- |
- |
Uncharacterized protein TCM_024268 [Theobroma cacao] |
| 2 |
Hb_001793_040 |
0.1668589533 |
- |
- |
hypothetical protein VITISV_006810 [Vitis vinifera] |
| 3 |
Hb_001385_070 |
0.1708435811 |
- |
- |
hypothetical protein VITISV_019194 [Vitis vinifera] |
| 4 |
Hb_170878_020 |
0.173729241 |
- |
- |
- |
| 5 |
Hb_008055_030 |
0.1767066769 |
- |
- |
hypothetical protein PRUPE_ppa022673mg [Prunus persica] |
| 6 |
Hb_127552_050 |
0.178604623 |
- |
- |
hypothetical protein VITISV_030752 [Vitis vinifera] |
| 7 |
Hb_000968_070 |
0.180476987 |
- |
- |
- |
| 8 |
Hb_000644_020 |
0.1828152184 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA1 [Jatropha curcas] |
| 9 |
Hb_007793_030 |
0.1862855223 |
- |
- |
OSJNBa0074B10.6 [Oryza sativa Japonica Group] |
| 10 |
Hb_005920_030 |
0.1887251642 |
- |
- |
putative polyprotein [Oryza sativa Japonica Group] |
| 11 |
Hb_182651_020 |
0.1901420997 |
- |
- |
alcohol dehydrogenase, putative [Ricinus communis] |
| 12 |
Hb_040473_010 |
0.1912117858 |
- |
- |
hypothetical protein POPTR_0001s08730g [Populus trichocarpa] |
| 13 |
Hb_000473_160 |
0.1919822083 |
- |
- |
PREDICTED: uncharacterized protein LOC102596609 [Solanum tuberosum] |
| 14 |
Hb_000574_330 |
0.1979663626 |
- |
- |
PREDICTED: clustered mitochondria protein homolog [Jatropha curcas] |
| 15 |
Hb_035651_010 |
0.1997690972 |
- |
- |
receptor-kinase, putative [Ricinus communis] |
| 16 |
Hb_011649_030 |
0.2006785983 |
- |
- |
hypothetical protein [Vitis hybrid cultivar] |
| 17 |
Hb_004520_010 |
0.2029860158 |
- |
- |
PREDICTED: uncharacterized protein LOC105631130 [Jatropha curcas] |
| 18 |
Hb_062013_010 |
0.2051659458 |
- |
- |
phosphoprotein phosphatase, putative [Ricinus communis] |
| 19 |
Hb_004021_020 |
0.2061818665 |
- |
- |
PREDICTED: transposon Tf2-1 polyprotein isoform X1 [Zea mays] |
| 20 |
Hb_002760_020 |
0.2077558685 |
- |
- |
PREDICTED: uncharacterized protein LOC105851424 [Cicer arietinum] |