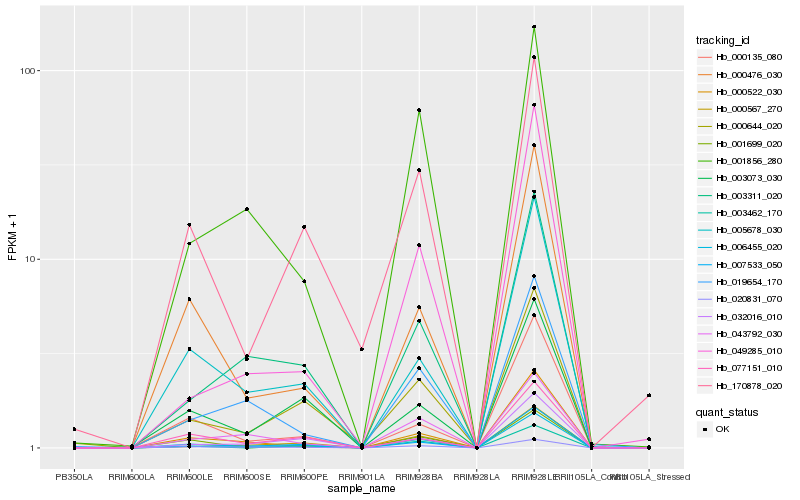
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006455_020 |
0.0 |
- |
- |
Ent-kaurene synthase B, chloroplast precursor, putative [Ricinus communis] |
| 2 |
Hb_020831_070 |
0.0839048327 |
- |
- |
hypothetical protein VITISV_027174 [Vitis vinifera] |
| 3 |
Hb_000476_030 |
0.1070308828 |
- |
- |
PREDICTED: uncharacterized protein LOC105629366 [Jatropha curcas] |
| 4 |
Hb_005678_030 |
0.1103254808 |
- |
- |
PREDICTED: uncharacterized protein At4g06744-like [Jatropha curcas] |
| 5 |
Hb_000644_020 |
0.1201945087 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA1 [Jatropha curcas] |
| 6 |
Hb_000522_030 |
0.1206543 |
- |
- |
hypothetical protein VITISV_006017 [Vitis vinifera] |
| 7 |
Hb_007533_050 |
0.120940372 |
- |
- |
- |
| 8 |
Hb_000567_270 |
0.1211141981 |
- |
- |
hypothetical protein L484_017162 [Morus notabilis] |
| 9 |
Hb_043792_030 |
0.1228639145 |
- |
- |
PREDICTED: uncharacterized protein LOC105632434 [Jatropha curcas] |
| 10 |
Hb_003311_020 |
0.1250122015 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000135_080 |
0.1275184369 |
- |
- |
Flavonol synthase/flavanone 3-hydroxylase, putative [Ricinus communis] |
| 12 |
Hb_001856_280 |
0.130076893 |
- |
- |
PREDICTED: receptor-like protein kinase FERONIA [Jatropha curcas] |
| 13 |
Hb_019654_170 |
0.1317457438 |
transcription factor |
TF Family: ARID |
PREDICTED: high mobility group B protein 9 [Jatropha curcas] |
| 14 |
Hb_003462_170 |
0.1326126644 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 15 |
Hb_032016_010 |
0.1340895042 |
- |
- |
PREDICTED: probably inactive leucine-rich repeat receptor-like protein kinase At3g28040-like isoform X1 [Citrus sinensis] |
| 16 |
Hb_170878_020 |
0.1388956085 |
- |
- |
- |
| 17 |
Hb_001699_020 |
0.141248746 |
- |
- |
hypothetical protein L484_026849 [Morus notabilis] |
| 18 |
Hb_077151_010 |
0.1444256899 |
- |
- |
ankyrin repeat protein [Bruguiera gymnorhiza] |
| 19 |
Hb_049285_010 |
0.1457391586 |
- |
- |
PREDICTED: terpene synthase 10-like [Jatropha curcas] |
| 20 |
Hb_003073_030 |
0.1457523021 |
- |
- |
conserved hypothetical protein [Ricinus communis] |