| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
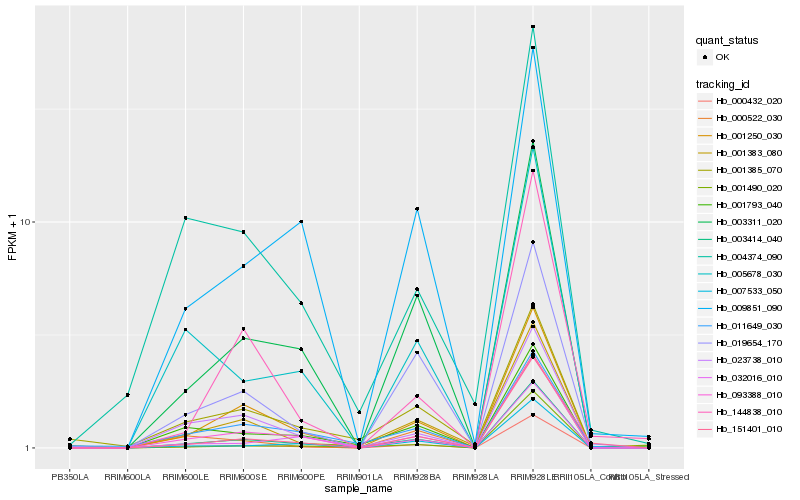
Hb_003414_040 |
0.0 |
- |
- |
hypothetical protein VITISV_016691 [Vitis vinifera] |
| 2 |
Hb_000432_020 |
0.0968536576 |
desease resistance |
Gene Name: Tropomyosin_1 |
PREDICTED: putative disease resistance protein RGA1 isoform X2 [Jatropha curcas] |
| 3 |
Hb_001383_080 |
0.1121704456 |
- |
- |
OSJNBa0019D11.8 [Oryza sativa Japonica Group] |
| 4 |
Hb_093388_010 |
0.1146983388 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO1 [Jatropha curcas] |
| 5 |
Hb_001250_030 |
0.1161228086 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_003311_020 |
0.1209696158 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_001793_040 |
0.1305164871 |
- |
- |
hypothetical protein VITISV_006810 [Vitis vinifera] |
| 8 |
Hb_007533_050 |
0.1340800345 |
- |
- |
- |
| 9 |
Hb_023738_010 |
0.1344127892 |
- |
- |
Concanavalin A-like lectin protein kinase family protein [Theobroma cacao] |
| 10 |
Hb_005678_030 |
0.1436699436 |
- |
- |
PREDICTED: uncharacterized protein At4g06744-like [Jatropha curcas] |
| 11 |
Hb_032016_010 |
0.1521221607 |
- |
- |
PREDICTED: probably inactive leucine-rich repeat receptor-like protein kinase At3g28040-like isoform X1 [Citrus sinensis] |
| 12 |
Hb_000522_030 |
0.1529987897 |
- |
- |
hypothetical protein VITISV_006017 [Vitis vinifera] |
| 13 |
Hb_151401_010 |
0.1538087758 |
- |
- |
- |
| 14 |
Hb_001490_020 |
0.1539322869 |
- |
- |
MULTISPECIES: hypothetical protein, partial [Pseudomonas fluorescens group] |
| 15 |
Hb_004374_090 |
0.1564415603 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox protein BEL1 homolog [Populus euphratica] |
| 16 |
Hb_011649_030 |
0.1578904179 |
- |
- |
hypothetical protein [Vitis hybrid cultivar] |
| 17 |
Hb_001385_070 |
0.1582833836 |
- |
- |
hypothetical protein VITISV_019194 [Vitis vinifera] |
| 18 |
Hb_019654_170 |
0.1601941203 |
transcription factor |
TF Family: ARID |
PREDICTED: high mobility group B protein 9 [Jatropha curcas] |
| 19 |
Hb_009851_090 |
0.1605065725 |
- |
- |
Protein ABIL2, putative [Ricinus communis] |
| 20 |
Hb_144838_010 |
0.1616369873 |
- |
- |
hypothetical protein JCGZ_08246 [Jatropha curcas] |