| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
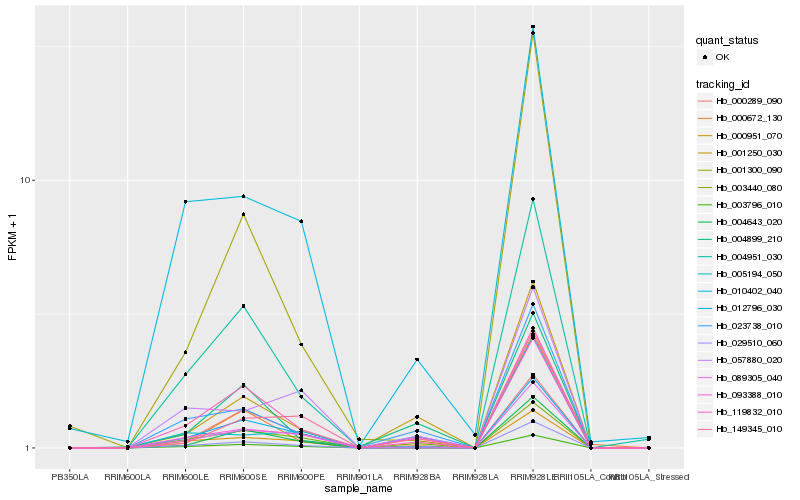
Hb_029510_060 |
0.0 |
- |
- |
PREDICTED: probable plastidic glucose transporter 3 [Jatropha curcas] |
| 2 |
Hb_023738_010 |
0.091164889 |
- |
- |
Concanavalin A-like lectin protein kinase family protein [Theobroma cacao] |
| 3 |
Hb_093388_010 |
0.094931017 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO1 [Jatropha curcas] |
| 4 |
Hb_119832_010 |
0.1062343994 |
- |
- |
- |
| 5 |
Hb_001250_030 |
0.113187424 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000951_070 |
0.1208585172 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: receptor-like protein kinase FERONIA [Populus euphratica] |
| 7 |
Hb_003796_010 |
0.125937251 |
- |
- |
PREDICTED: uncharacterized protein LOC105631113 [Jatropha curcas] |
| 8 |
Hb_003440_080 |
0.129944517 |
- |
- |
polyprotein [Citrus endogenous pararetrovirus] |
| 9 |
Hb_012796_030 |
0.1302108068 |
- |
- |
PREDICTED: uncharacterized protein LOC104106381 [Nicotiana tomentosiformis] |
| 10 |
Hb_149345_010 |
0.1397367699 |
- |
- |
Anthranilate N-benzoyltransferase protein, putative [Ricinus communis] |
| 11 |
Hb_089305_040 |
0.1415457988 |
- |
- |
- |
| 12 |
Hb_001300_090 |
0.1429152696 |
- |
- |
PREDICTED: glutaredoxin-C11 [Jatropha curcas] |
| 13 |
Hb_004951_030 |
0.1437485582 |
- |
- |
9-cis-epoxycarotenoid dioxygenase, putative [Ricinus communis] |
| 14 |
Hb_004643_020 |
0.1453174218 |
- |
- |
hypothetical protein AMTR_s05716p00004100, partial [Amborella trichopoda] |
| 15 |
Hb_010402_040 |
0.1462199261 |
- |
- |
hypothetical protein JCGZ_21227 [Jatropha curcas] |
| 16 |
Hb_000289_090 |
0.1525042712 |
- |
- |
- |
| 17 |
Hb_000672_130 |
0.1534910588 |
- |
- |
hypothetical protein POPTR_0010s22870g [Populus trichocarpa] |
| 18 |
Hb_004899_210 |
0.1540310073 |
- |
- |
PREDICTED: respiratory burst oxidase homolog protein E [Jatropha curcas] |
| 19 |
Hb_005194_050 |
0.1552418788 |
transcription factor |
TF Family: GRAS |
SCL domain class transcription factor [Theobroma cacao] |
| 20 |
Hb_057880_020 |
0.1554899489 |
- |
- |
PREDICTED: receptor-like protein kinase HSL1, partial [Jatropha curcas] |