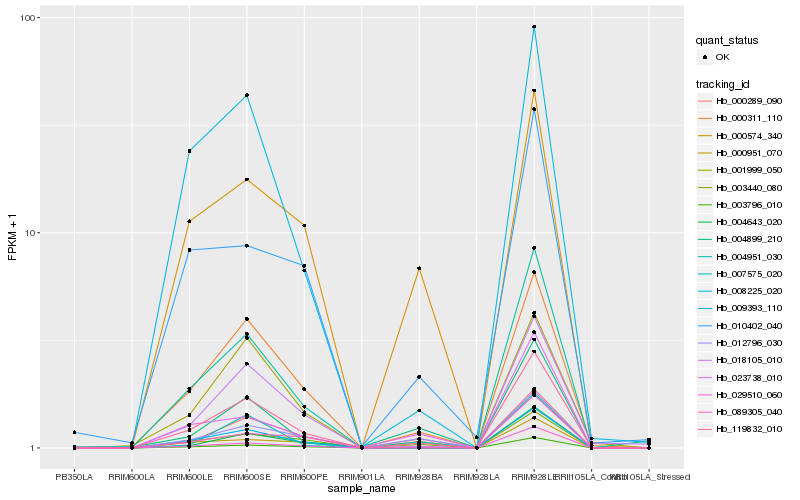
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_119832_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_000311_110 |
0.0877359876 |
- |
- |
ATP-binding cassette transporter, putative [Ricinus communis] |
| 3 |
Hb_009393_110 |
0.1004883573 |
- |
- |
PREDICTED: circadian locomoter output cycles protein kaput [Jatropha curcas] |
| 4 |
Hb_029510_060 |
0.1062343994 |
- |
- |
PREDICTED: probable plastidic glucose transporter 3 [Jatropha curcas] |
| 5 |
Hb_004951_030 |
0.1101540752 |
- |
- |
9-cis-epoxycarotenoid dioxygenase, putative [Ricinus communis] |
| 6 |
Hb_003440_080 |
0.1148952401 |
- |
- |
polyprotein [Citrus endogenous pararetrovirus] |
| 7 |
Hb_003796_010 |
0.1168624623 |
- |
- |
PREDICTED: uncharacterized protein LOC105631113 [Jatropha curcas] |
| 8 |
Hb_000951_070 |
0.1198592428 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: receptor-like protein kinase FERONIA [Populus euphratica] |
| 9 |
Hb_000289_090 |
0.1267743793 |
- |
- |
- |
| 10 |
Hb_008225_020 |
0.1292117292 |
- |
- |
PREDICTED: cytochrome P450 71A26-like [Jatropha curcas] |
| 11 |
Hb_018105_010 |
0.129440284 |
- |
- |
hypothetical protein POPTR_0004s08710g [Populus trichocarpa] |
| 12 |
Hb_012796_030 |
0.1352440588 |
- |
- |
PREDICTED: uncharacterized protein LOC104106381 [Nicotiana tomentosiformis] |
| 13 |
Hb_001999_050 |
0.135487724 |
rubber biosynthesis |
Gene Name: CPT7 |
PREDICTED: dehydrodolichyl diphosphate synthase 2 [Jatropha curcas] |
| 14 |
Hb_004643_020 |
0.1357831536 |
- |
- |
hypothetical protein AMTR_s05716p00004100, partial [Amborella trichopoda] |
| 15 |
Hb_089305_040 |
0.1555958121 |
- |
- |
- |
| 16 |
Hb_010402_040 |
0.1603629014 |
- |
- |
hypothetical protein JCGZ_21227 [Jatropha curcas] |
| 17 |
Hb_007575_020 |
0.1604312356 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 18 |
Hb_023738_010 |
0.1633040338 |
- |
- |
Concanavalin A-like lectin protein kinase family protein [Theobroma cacao] |
| 19 |
Hb_000574_340 |
0.1633610945 |
- |
- |
hypothetical protein RCOM_0901420 [Ricinus communis] |
| 20 |
Hb_004899_210 |
0.1646272728 |
- |
- |
PREDICTED: respiratory burst oxidase homolog protein E [Jatropha curcas] |