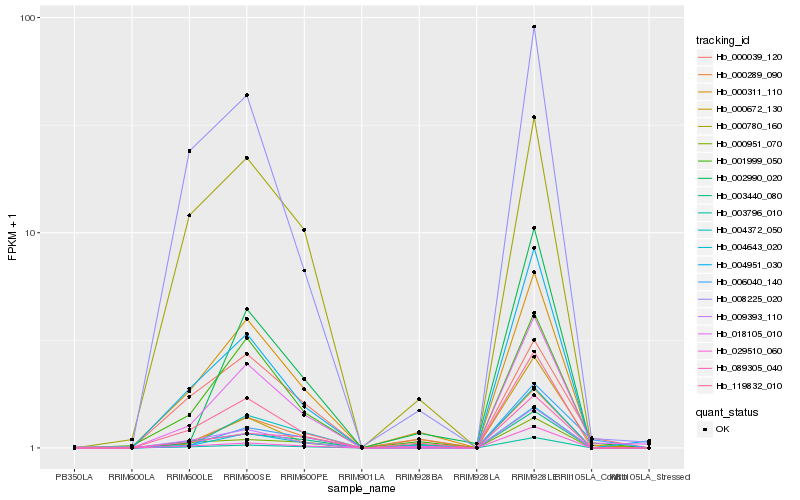
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_018105_010 |
0.0 |
- |
- |
hypothetical protein POPTR_0004s08710g [Populus trichocarpa] |
| 2 |
Hb_001999_050 |
0.1146749129 |
rubber biosynthesis |
Gene Name: CPT7 |
PREDICTED: dehydrodolichyl diphosphate synthase 2 [Jatropha curcas] |
| 3 |
Hb_000311_110 |
0.1180243038 |
- |
- |
ATP-binding cassette transporter, putative [Ricinus communis] |
| 4 |
Hb_004951_030 |
0.124274074 |
- |
- |
9-cis-epoxycarotenoid dioxygenase, putative [Ricinus communis] |
| 5 |
Hb_009393_110 |
0.1284149257 |
- |
- |
PREDICTED: circadian locomoter output cycles protein kaput [Jatropha curcas] |
| 6 |
Hb_119832_010 |
0.129440284 |
- |
- |
- |
| 7 |
Hb_004643_020 |
0.1390592199 |
- |
- |
hypothetical protein AMTR_s05716p00004100, partial [Amborella trichopoda] |
| 8 |
Hb_003796_010 |
0.145931047 |
- |
- |
PREDICTED: uncharacterized protein LOC105631113 [Jatropha curcas] |
| 9 |
Hb_004372_050 |
0.1631407579 |
- |
- |
putative receptor-like protein kinase [Morus notabilis] |
| 10 |
Hb_089305_040 |
0.1696453769 |
- |
- |
- |
| 11 |
Hb_008225_020 |
0.172335566 |
- |
- |
PREDICTED: cytochrome P450 71A26-like [Jatropha curcas] |
| 12 |
Hb_000951_070 |
0.1756667949 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: receptor-like protein kinase FERONIA [Populus euphratica] |
| 13 |
Hb_002990_020 |
0.1813583861 |
- |
- |
PREDICTED: vacuolar amino acid transporter 1 [Jatropha curcas] |
| 14 |
Hb_000289_090 |
0.1851497575 |
- |
- |
- |
| 15 |
Hb_003440_080 |
0.186831337 |
- |
- |
polyprotein [Citrus endogenous pararetrovirus] |
| 16 |
Hb_029510_060 |
0.1899309887 |
- |
- |
PREDICTED: probable plastidic glucose transporter 3 [Jatropha curcas] |
| 17 |
Hb_000672_130 |
0.1936056647 |
- |
- |
hypothetical protein POPTR_0010s22870g [Populus trichocarpa] |
| 18 |
Hb_006040_140 |
0.1945432909 |
- |
- |
hypothetical protein NitaMp105 [Nicotiana tabacum] |
| 19 |
Hb_000780_160 |
0.194684441 |
- |
- |
hypothetical protein RCOM_0483300 [Ricinus communis] |
| 20 |
Hb_000039_120 |
0.1947385473 |
- |
- |
PREDICTED: equilibrative nucleotide transporter 3-like [Jatropha curcas] |