| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
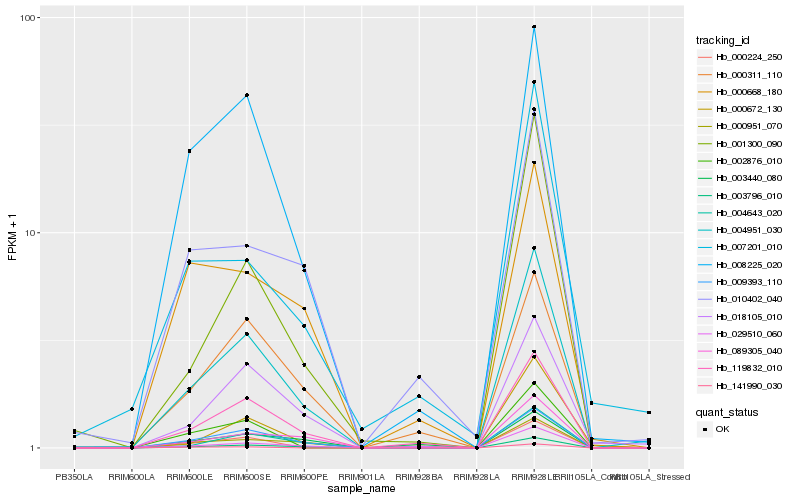
Hb_004951_030 |
0.0 |
- |
- |
9-cis-epoxycarotenoid dioxygenase, putative [Ricinus communis] |
| 2 |
Hb_003796_010 |
0.0821259269 |
- |
- |
PREDICTED: uncharacterized protein LOC105631113 [Jatropha curcas] |
| 3 |
Hb_009393_110 |
0.0848929782 |
- |
- |
PREDICTED: circadian locomoter output cycles protein kaput [Jatropha curcas] |
| 4 |
Hb_119832_010 |
0.1101540752 |
- |
- |
- |
| 5 |
Hb_004643_020 |
0.1182942423 |
- |
- |
hypothetical protein AMTR_s05716p00004100, partial [Amborella trichopoda] |
| 6 |
Hb_018105_010 |
0.124274074 |
- |
- |
hypothetical protein POPTR_0004s08710g [Populus trichocarpa] |
| 7 |
Hb_008225_020 |
0.1290453442 |
- |
- |
PREDICTED: cytochrome P450 71A26-like [Jatropha curcas] |
| 8 |
Hb_089305_040 |
0.132476448 |
- |
- |
- |
| 9 |
Hb_000951_070 |
0.137122089 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: receptor-like protein kinase FERONIA [Populus euphratica] |
| 10 |
Hb_029510_060 |
0.1437485582 |
- |
- |
PREDICTED: probable plastidic glucose transporter 3 [Jatropha curcas] |
| 11 |
Hb_001300_090 |
0.1550823558 |
- |
- |
PREDICTED: glutaredoxin-C11 [Jatropha curcas] |
| 12 |
Hb_000311_110 |
0.1584828753 |
- |
- |
ATP-binding cassette transporter, putative [Ricinus communis] |
| 13 |
Hb_000224_250 |
0.1716958874 |
- |
- |
hypothetical protein AMTR_s05400p00003230 [Amborella trichopoda] |
| 14 |
Hb_010402_040 |
0.1751322957 |
- |
- |
hypothetical protein JCGZ_21227 [Jatropha curcas] |
| 15 |
Hb_002876_010 |
0.1752574876 |
transcription factor |
TF Family: MYB-related |
DNA binding protein, putative [Ricinus communis] |
| 16 |
Hb_000672_130 |
0.1823456487 |
- |
- |
hypothetical protein POPTR_0010s22870g [Populus trichocarpa] |
| 17 |
Hb_000668_180 |
0.1825955996 |
- |
- |
hypothetical protein POPTR_0005s16490g [Populus trichocarpa] |
| 18 |
Hb_007201_010 |
0.1843812314 |
- |
- |
PREDICTED: uncharacterized protein LOC105631226 isoform X2 [Jatropha curcas] |
| 19 |
Hb_141990_030 |
0.1865863088 |
- |
- |
hypothetical protein JCGZ_09181 [Jatropha curcas] |
| 20 |
Hb_003440_080 |
0.1867936207 |
- |
- |
polyprotein [Citrus endogenous pararetrovirus] |