| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
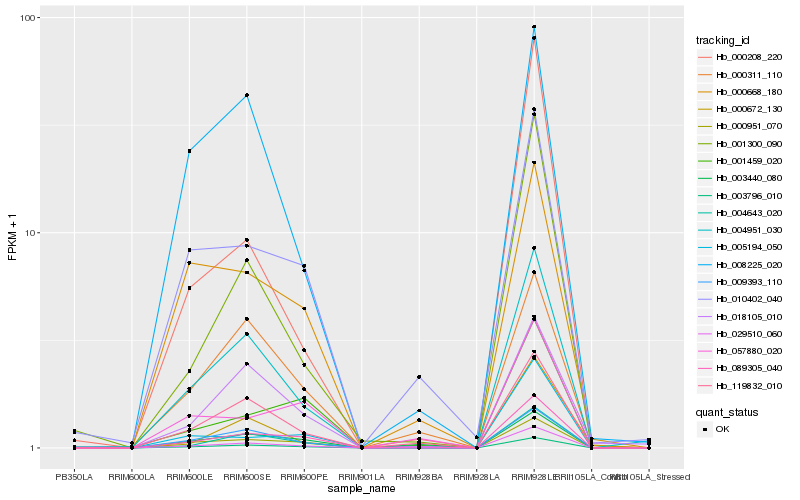
Hb_003796_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105631113 [Jatropha curcas] |
| 2 |
Hb_089305_040 |
0.0638999204 |
- |
- |
- |
| 3 |
Hb_004643_020 |
0.0820692474 |
- |
- |
hypothetical protein AMTR_s05716p00004100, partial [Amborella trichopoda] |
| 4 |
Hb_004951_030 |
0.0821259269 |
- |
- |
9-cis-epoxycarotenoid dioxygenase, putative [Ricinus communis] |
| 5 |
Hb_009393_110 |
0.0829213999 |
- |
- |
PREDICTED: circadian locomoter output cycles protein kaput [Jatropha curcas] |
| 6 |
Hb_000951_070 |
0.1040539112 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: receptor-like protein kinase FERONIA [Populus euphratica] |
| 7 |
Hb_119832_010 |
0.1168624623 |
- |
- |
- |
| 8 |
Hb_029510_060 |
0.125937251 |
- |
- |
PREDICTED: probable plastidic glucose transporter 3 [Jatropha curcas] |
| 9 |
Hb_001300_090 |
0.140013279 |
- |
- |
PREDICTED: glutaredoxin-C11 [Jatropha curcas] |
| 10 |
Hb_018105_010 |
0.145931047 |
- |
- |
hypothetical protein POPTR_0004s08710g [Populus trichocarpa] |
| 11 |
Hb_010402_040 |
0.1601302938 |
- |
- |
hypothetical protein JCGZ_21227 [Jatropha curcas] |
| 12 |
Hb_000311_110 |
0.1625048543 |
- |
- |
ATP-binding cassette transporter, putative [Ricinus communis] |
| 13 |
Hb_008225_020 |
0.1636970336 |
- |
- |
PREDICTED: cytochrome P450 71A26-like [Jatropha curcas] |
| 14 |
Hb_001459_020 |
0.1665693738 |
- |
- |
ankyrin repeat protein [Bruguiera gymnorhiza] |
| 15 |
Hb_003440_080 |
0.170775758 |
- |
- |
polyprotein [Citrus endogenous pararetrovirus] |
| 16 |
Hb_000668_180 |
0.1712750896 |
- |
- |
hypothetical protein POPTR_0005s16490g [Populus trichocarpa] |
| 17 |
Hb_005194_050 |
0.1732783916 |
transcription factor |
TF Family: GRAS |
SCL domain class transcription factor [Theobroma cacao] |
| 18 |
Hb_000672_130 |
0.1805118466 |
- |
- |
hypothetical protein POPTR_0010s22870g [Populus trichocarpa] |
| 19 |
Hb_000208_220 |
0.1807315934 |
- |
- |
- |
| 20 |
Hb_057880_020 |
0.1864712737 |
- |
- |
PREDICTED: receptor-like protein kinase HSL1, partial [Jatropha curcas] |