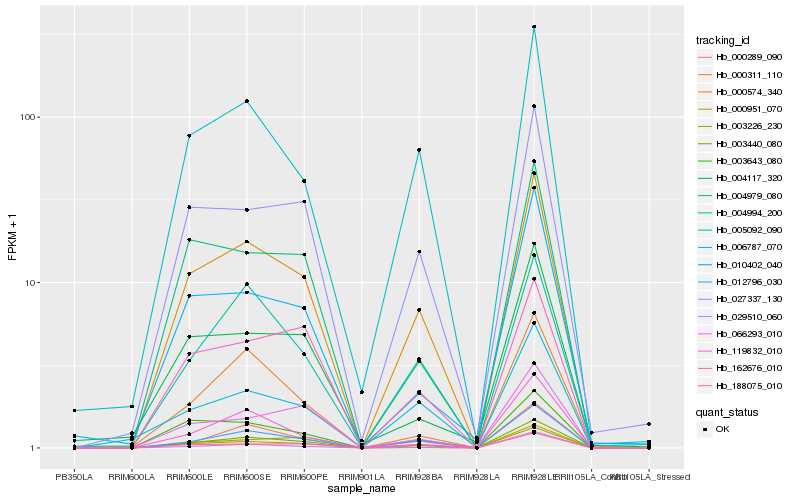
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003440_080 |
0.0 |
- |
- |
polyprotein [Citrus endogenous pararetrovirus] |
| 2 |
Hb_000574_340 |
0.0660869814 |
- |
- |
hypothetical protein RCOM_0901420 [Ricinus communis] |
| 3 |
Hb_012796_030 |
0.0679403159 |
- |
- |
PREDICTED: uncharacterized protein LOC104106381 [Nicotiana tomentosiformis] |
| 4 |
Hb_000289_090 |
0.1080904621 |
- |
- |
- |
| 5 |
Hb_000951_070 |
0.1083139098 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: receptor-like protein kinase FERONIA [Populus euphratica] |
| 6 |
Hb_119832_010 |
0.1148952401 |
- |
- |
- |
| 7 |
Hb_027337_130 |
0.1187140666 |
- |
- |
PREDICTED: non-functional NADPH-dependent codeinone reductase 2-like [Jatropha curcas] |
| 8 |
Hb_000311_110 |
0.1230148245 |
- |
- |
ATP-binding cassette transporter, putative [Ricinus communis] |
| 9 |
Hb_010402_040 |
0.124482981 |
- |
- |
hypothetical protein JCGZ_21227 [Jatropha curcas] |
| 10 |
Hb_005092_090 |
0.1251211571 |
- |
- |
cold regulated protein [Manihot esculenta] |
| 11 |
Hb_066293_010 |
0.126171422 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g35710 [Jatropha curcas] |
| 12 |
Hb_029510_060 |
0.129944517 |
- |
- |
PREDICTED: probable plastidic glucose transporter 3 [Jatropha curcas] |
| 13 |
Hb_004979_080 |
0.1309033368 |
- |
- |
kinase, putative [Ricinus communis] |
| 14 |
Hb_003643_080 |
0.1321890081 |
- |
- |
hypothetical protein JCGZ_01308 [Jatropha curcas] |
| 15 |
Hb_188075_010 |
0.1324429158 |
- |
- |
PREDICTED: uncharacterized protein LOC105775124 [Gossypium raimondii] |
| 16 |
Hb_004994_200 |
0.1347448531 |
- |
- |
PREDICTED: uncharacterized protein LOC105174340 [Sesamum indicum] |
| 17 |
Hb_006787_070 |
0.1400818236 |
- |
- |
PREDICTED: uncharacterized protein LOC104121706, partial [Nicotiana tomentosiformis] |
| 18 |
Hb_004117_320 |
0.1402996735 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase WNK5 [Jatropha curcas] |
| 19 |
Hb_003226_230 |
0.143613234 |
- |
- |
PREDICTED: transposon Ty3-G Gag-Pol polyprotein [Phoenix dactylifera] |
| 20 |
Hb_162676_010 |
0.1437635861 |
- |
- |
ATP binding protein, putative [Ricinus communis] |