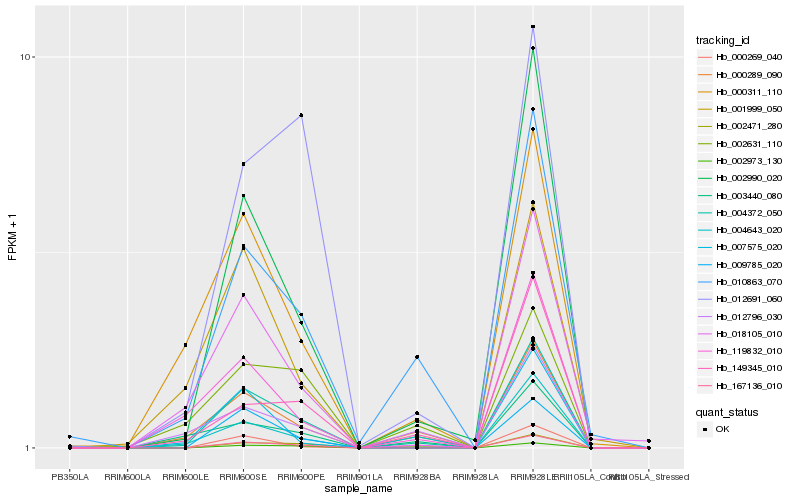
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004372_050 |
0.0 |
- |
- |
putative receptor-like protein kinase [Morus notabilis] |
| 2 |
Hb_002990_020 |
0.0940349104 |
- |
- |
PREDICTED: vacuolar amino acid transporter 1 [Jatropha curcas] |
| 3 |
Hb_167136_010 |
0.1214620028 |
- |
- |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 4 |
Hb_000289_090 |
0.1359358511 |
- |
- |
- |
| 5 |
Hb_010863_070 |
0.1466574233 |
- |
- |
PREDICTED: glutaredoxin-C11 [Jatropha curcas] |
| 6 |
Hb_002471_280 |
0.147879956 |
- |
- |
PREDICTED: cyclin-SDS [Jatropha curcas] |
| 7 |
Hb_004643_020 |
0.1499106135 |
- |
- |
hypothetical protein AMTR_s05716p00004100, partial [Amborella trichopoda] |
| 8 |
Hb_018105_010 |
0.1631407579 |
- |
- |
hypothetical protein POPTR_0004s08710g [Populus trichocarpa] |
| 9 |
Hb_009785_020 |
0.1644220641 |
- |
- |
PREDICTED: nucleolin [Populus euphratica] |
| 10 |
Hb_119832_010 |
0.164644084 |
- |
- |
- |
| 11 |
Hb_000311_110 |
0.1662099023 |
- |
- |
ATP-binding cassette transporter, putative [Ricinus communis] |
| 12 |
Hb_149345_010 |
0.1686161236 |
- |
- |
Anthranilate N-benzoyltransferase protein, putative [Ricinus communis] |
| 13 |
Hb_001999_050 |
0.1811903627 |
rubber biosynthesis |
Gene Name: CPT7 |
PREDICTED: dehydrodolichyl diphosphate synthase 2 [Jatropha curcas] |
| 14 |
Hb_012691_060 |
0.1814840808 |
- |
- |
PREDICTED: isoflavone 2'-hydroxylase-like isoform X1 [Prunus mume] |
| 15 |
Hb_012796_030 |
0.1828315056 |
- |
- |
PREDICTED: uncharacterized protein LOC104106381 [Nicotiana tomentosiformis] |
| 16 |
Hb_002973_130 |
0.1831540025 |
- |
- |
PREDICTED: uncharacterized protein LOC104609908 [Nelumbo nucifera] |
| 17 |
Hb_000269_040 |
0.1832340545 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 18 |
Hb_003440_080 |
0.1848044921 |
- |
- |
polyprotein [Citrus endogenous pararetrovirus] |
| 19 |
Hb_007575_020 |
0.1858849201 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 20 |
Hb_002631_110 |
0.190740405 |
- |
- |
PREDICTED: cytochrome P450 71D9-like [Fragaria vesca subsp. vesca] |