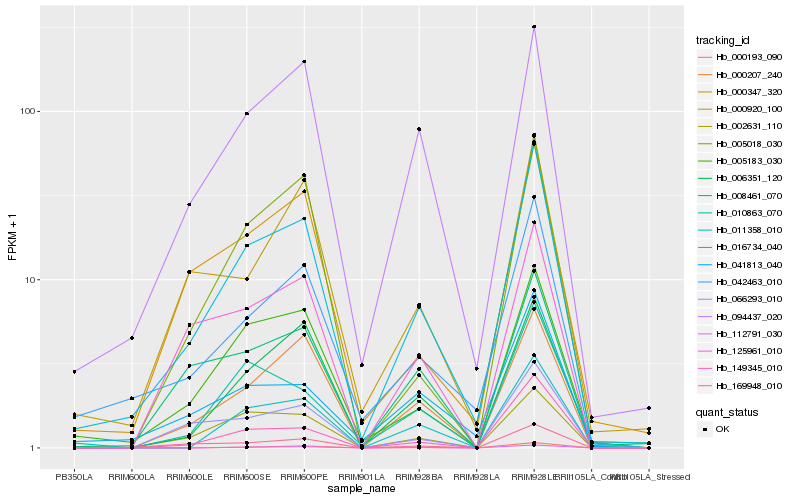
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_016734_040 |
0.0 |
- |
- |
abhydrolase domain containing, putative [Ricinus communis] |
| 2 |
Hb_042463_010 |
0.1120278578 |
- |
- |
cytochrome c oxidase subunit 2-1 (mitochondrion) (mitochondrion) [Brassica juncea var. tumida] |
| 3 |
Hb_005183_030 |
0.1235403314 |
- |
- |
PREDICTED: probable calcium-binding protein CML16 [Jatropha curcas] |
| 4 |
Hb_169948_010 |
0.1351462679 |
- |
- |
hypothetical protein VITISV_006524 [Vitis vinifera] |
| 5 |
Hb_094437_020 |
0.1366219499 |
- |
- |
Xylem serine proteinase 1 precursor, putative [Ricinus communis] |
| 6 |
Hb_149345_010 |
0.1389259489 |
- |
- |
Anthranilate N-benzoyltransferase protein, putative [Ricinus communis] |
| 7 |
Hb_002631_110 |
0.1423187524 |
- |
- |
PREDICTED: cytochrome P450 71D9-like [Fragaria vesca subsp. vesca] |
| 8 |
Hb_000347_320 |
0.1455861056 |
- |
- |
PREDICTED: subtilisin-like protease SBT1.7 [Jatropha curcas] |
| 9 |
Hb_000207_240 |
0.146804725 |
- |
- |
hypothetical protein RCOM_0686540 [Ricinus communis] |
| 10 |
Hb_041813_040 |
0.1475700364 |
- |
- |
NADH2 dehydrogenase (ubiquinone) (EC 1.6.5.3) chain 5, truncated - sugar beet mitochondrion (mitochondrion) [Beta vulgaris subsp. vulgaris] |
| 11 |
Hb_006351_120 |
0.1531155783 |
- |
- |
hypothetical protein CICLE_v10030803mg [Citrus clementina] |
| 12 |
Hb_066293_010 |
0.1543109754 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g35710 [Jatropha curcas] |
| 13 |
Hb_000920_100 |
0.1547308871 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 14 |
Hb_011358_010 |
0.1550011197 |
- |
- |
- |
| 15 |
Hb_010863_070 |
0.1558308772 |
- |
- |
PREDICTED: glutaredoxin-C11 [Jatropha curcas] |
| 16 |
Hb_008461_070 |
0.1561949799 |
- |
- |
PREDICTED: ethylene-overproduction protein 1 isoform X1 [Jatropha curcas] |
| 17 |
Hb_005018_030 |
0.1566397724 |
- |
- |
hypothetical protein CICLE_v10019811mg [Citrus clementina] |
| 18 |
Hb_125961_010 |
0.1613325008 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 19 |
Hb_000193_090 |
0.1623118404 |
- |
- |
cysteine protease, putative [Ricinus communis] |
| 20 |
Hb_112791_030 |
0.1660315625 |
- |
- |
Uncharacterized protein TCM_027940 [Theobroma cacao] |