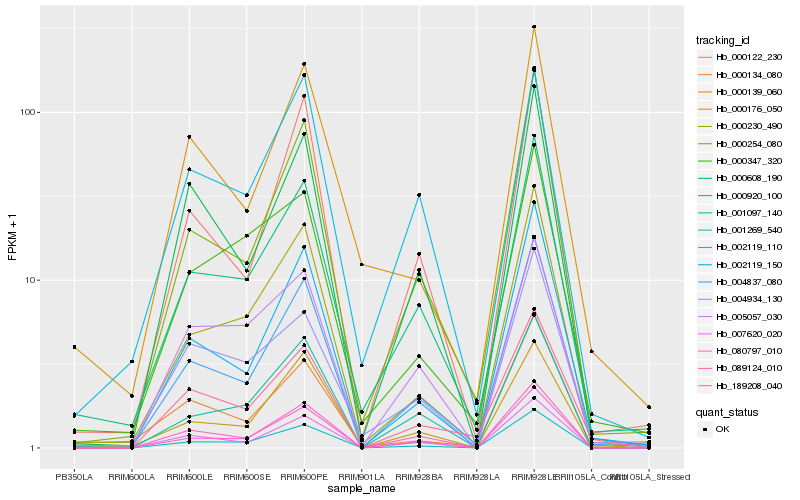
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000920_100 |
0.0 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 2 |
Hb_004837_080 |
0.0848829406 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB46 [Jatropha curcas] |
| 3 |
Hb_000254_080 |
0.0938390429 |
- |
- |
PREDICTED: uncharacterized protein LOC105635000 [Jatropha curcas] |
| 4 |
Hb_000230_490 |
0.0980684776 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_004934_130 |
0.1050700527 |
- |
- |
12-oxophytodienoate reductase 2 isoform 1 [Theobroma cacao] |
| 6 |
Hb_089124_010 |
0.1082156226 |
- |
- |
hypothetical protein POPTR_0018s09540g [Populus trichocarpa] |
| 7 |
Hb_001097_140 |
0.1092979259 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_001269_540 |
0.1106671698 |
- |
- |
hypothetical protein VITISV_038016 [Vitis vinifera] |
| 9 |
Hb_007620_020 |
0.1132064162 |
- |
- |
PREDICTED: cationic amino acid transporter 7, chloroplastic-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_080797_010 |
0.1146798555 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At3g47570-like [Citrus sinensis] |
| 11 |
Hb_000347_320 |
0.1175878558 |
- |
- |
PREDICTED: subtilisin-like protease SBT1.7 [Jatropha curcas] |
| 12 |
Hb_000122_230 |
0.1186472588 |
- |
- |
laccase, putative [Ricinus communis] |
| 13 |
Hb_000176_050 |
0.120067942 |
- |
- |
glutamine amidotransferase class-I domain-containing family protein [Populus trichocarpa] |
| 14 |
Hb_000134_080 |
0.122221552 |
- |
- |
synaptotagmin, putative [Ricinus communis] |
| 15 |
Hb_000139_060 |
0.1229477236 |
- |
- |
Photosystem I reaction center subunit II, chloroplast precursor, putative [Ricinus communis] |
| 16 |
Hb_002119_150 |
0.1236139787 |
- |
- |
hypothetical protein POPTR_0008s13870g [Populus trichocarpa] |
| 17 |
Hb_000608_190 |
0.124570089 |
rubber biosynthesis |
Gene Name: Isopentenyl-diphosphate delta isomerase |
PREDICTED: isopentenyl-diphosphate Delta-isomerase I [Jatropha curcas] |
| 18 |
Hb_002119_110 |
0.1259992763 |
- |
- |
PREDICTED: probable xyloglucan endotransglucosylase/hydrolase protein 28 [Jatropha curcas] |
| 19 |
Hb_005057_030 |
0.1280860981 |
- |
- |
Leucoanthocyanidin dioxygenase, putative [Ricinus communis] |
| 20 |
Hb_189208_040 |
0.1291000181 |
- |
- |
hypothetical protein POPTR_0002s18920g [Populus trichocarpa] |