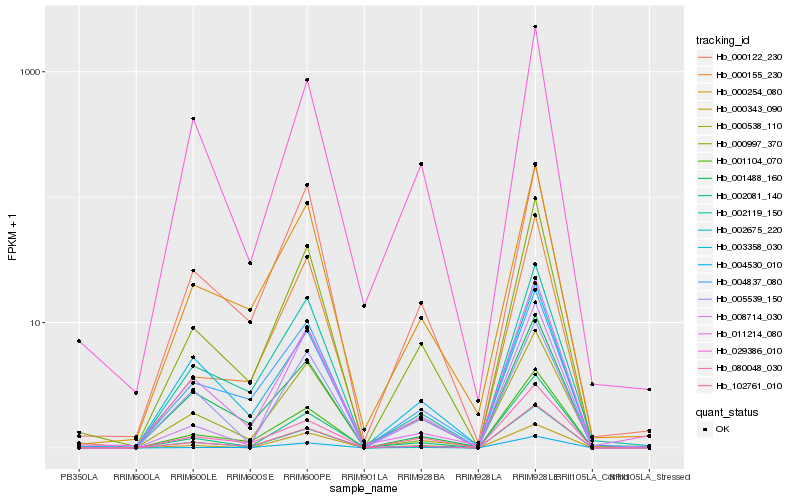
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000997_370 |
0.0 |
- |
- |
PREDICTED: 3-ketoacyl-CoA synthase 11-like [Jatropha curcas] |
| 2 |
Hb_002081_140 |
0.0875659436 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000254_080 |
0.0900938472 |
- |
- |
PREDICTED: uncharacterized protein LOC105635000 [Jatropha curcas] |
| 4 |
Hb_002119_150 |
0.1075250606 |
- |
- |
hypothetical protein POPTR_0008s13870g [Populus trichocarpa] |
| 5 |
Hb_004837_080 |
0.1096005276 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB46 [Jatropha curcas] |
| 6 |
Hb_000538_110 |
0.1098215004 |
- |
- |
sucrose synthase 1 [Hevea brasiliensis] |
| 7 |
Hb_029386_010 |
0.1108542709 |
- |
- |
RALF-LIKE 27 family protein [Populus trichocarpa] |
| 8 |
Hb_002675_220 |
0.1130603002 |
- |
- |
PREDICTED: probable pyridoxal biosynthesis protein PDX1.2 [Populus euphratica] |
| 9 |
Hb_000122_230 |
0.114927323 |
- |
- |
laccase, putative [Ricinus communis] |
| 10 |
Hb_102761_010 |
0.1149317494 |
- |
- |
hypothetical protein JCGZ_21614 [Jatropha curcas] |
| 11 |
Hb_001104_070 |
0.1150249914 |
- |
- |
hypothetical protein SORBIDRAFT_10g024640 [Sorghum bicolor] |
| 12 |
Hb_000343_090 |
0.1183741994 |
transcription factor |
TF Family: OFP |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_004530_010 |
0.1193454045 |
- |
- |
PREDICTED: uncharacterized protein LOC104898079 [Beta vulgaris subsp. vulgaris] |
| 14 |
Hb_011214_080 |
0.1214147159 |
- |
- |
sucrose synthase 1 [Hevea brasiliensis] |
| 15 |
Hb_080048_030 |
0.1214817934 |
desease resistance |
Gene Name: NB-ARC |
Leucine-rich repeat containing protein [Theobroma cacao] |
| 16 |
Hb_000155_230 |
0.1218206566 |
- |
- |
rac gtpase, putative [Ricinus communis] |
| 17 |
Hb_003358_030 |
0.1218273944 |
- |
- |
ankyrin-kinase, putative [Ricinus communis] |
| 18 |
Hb_001488_160 |
0.1241337679 |
- |
- |
UDP-glucuronosyltransferase, putative [Ricinus communis] |
| 19 |
Hb_005539_150 |
0.1244084756 |
- |
- |
PREDICTED: probable xyloglucan endotransglucosylase/hydrolase protein 33 [Jatropha curcas] |
| 20 |
Hb_008714_030 |
0.1252101283 |
- |
- |
Multidrug resistance protein ABC transporter family protein, putative [Theobroma cacao] |