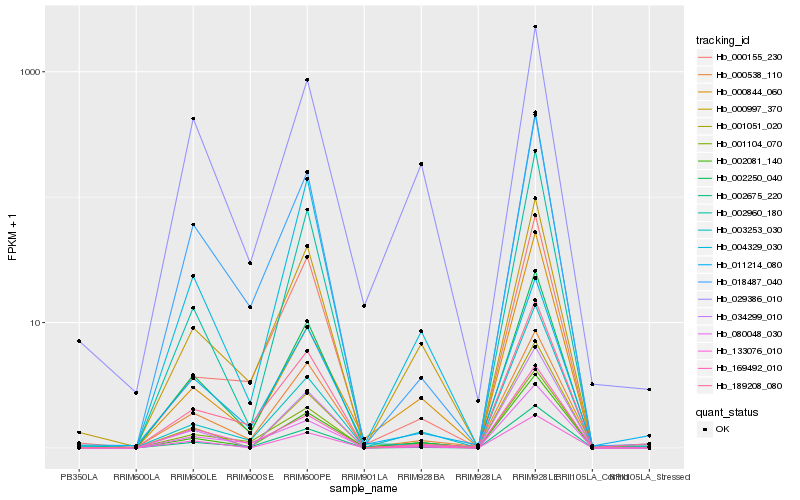
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002081_140 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_004329_030 |
0.0729904201 |
- |
- |
hypothetical protein JCGZ_17335 [Jatropha curcas] |
| 3 |
Hb_001051_020 |
0.0855304603 |
- |
- |
hypothetical protein L484_008044 [Morus notabilis] |
| 4 |
Hb_000997_370 |
0.0875659436 |
- |
- |
PREDICTED: 3-ketoacyl-CoA synthase 11-like [Jatropha curcas] |
| 5 |
Hb_002675_220 |
0.0938180534 |
- |
- |
PREDICTED: probable pyridoxal biosynthesis protein PDX1.2 [Populus euphratica] |
| 6 |
Hb_003253_030 |
0.0951029993 |
- |
- |
lysosomal pro-X carboxypeptidase, putative [Ricinus communis] |
| 7 |
Hb_034299_010 |
0.0982403128 |
- |
- |
F-box and wd40 domain protein, putative [Ricinus communis] |
| 8 |
Hb_080048_030 |
0.1029407175 |
desease resistance |
Gene Name: NB-ARC |
Leucine-rich repeat containing protein [Theobroma cacao] |
| 9 |
Hb_001104_070 |
0.1077525711 |
- |
- |
hypothetical protein SORBIDRAFT_10g024640 [Sorghum bicolor] |
| 10 |
Hb_169492_010 |
0.1124350312 |
- |
- |
PREDICTED: expansin-like B1 [Jatropha curcas] |
| 11 |
Hb_002250_040 |
0.1131940122 |
- |
- |
PREDICTED: transcription repressor OFP17 [Jatropha curcas] |
| 12 |
Hb_000844_060 |
0.1169986865 |
- |
- |
PREDICTED: uncharacterized protein LOC105633706 [Jatropha curcas] |
| 13 |
Hb_011214_080 |
0.1177450134 |
- |
- |
sucrose synthase 1 [Hevea brasiliensis] |
| 14 |
Hb_018487_040 |
0.1207520566 |
- |
- |
PREDICTED: 3-ketoacyl-CoA synthase 6 [Jatropha curcas] |
| 15 |
Hb_002960_180 |
0.1231171417 |
- |
- |
PREDICTED: GDSL esterase/lipase EXL3-like [Jatropha curcas] |
| 16 |
Hb_000538_110 |
0.1239874376 |
- |
- |
sucrose synthase 1 [Hevea brasiliensis] |
| 17 |
Hb_000155_230 |
0.1253198743 |
- |
- |
rac gtpase, putative [Ricinus communis] |
| 18 |
Hb_189208_080 |
0.1253955916 |
desease resistance |
Gene Name: ABC_membrane |
PREDICTED: ABC transporter B family member 11-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_133076_010 |
0.1271032677 |
- |
- |
PREDICTED: scopoletin glucosyltransferase-like [Populus euphratica] |
| 20 |
Hb_029386_010 |
0.128527443 |
- |
- |
RALF-LIKE 27 family protein [Populus trichocarpa] |