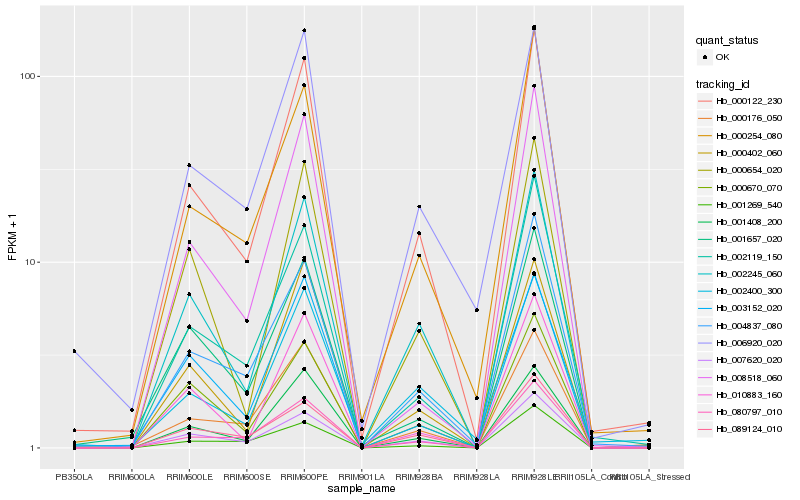
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000122_230 |
0.0 |
- |
- |
laccase, putative [Ricinus communis] |
| 2 |
Hb_002245_060 |
0.0706094009 |
- |
- |
PREDICTED: kinesin-3 isoform X1 [Jatropha curcas] |
| 3 |
Hb_004837_080 |
0.0728384166 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB46 [Jatropha curcas] |
| 4 |
Hb_001408_200 |
0.0759044935 |
- |
- |
hypothetical protein JCGZ_08964 [Jatropha curcas] |
| 5 |
Hb_080797_010 |
0.0791861144 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At3g47570-like [Citrus sinensis] |
| 6 |
Hb_000254_080 |
0.0831566619 |
- |
- |
PREDICTED: uncharacterized protein LOC105635000 [Jatropha curcas] |
| 7 |
Hb_007620_020 |
0.0841632256 |
- |
- |
PREDICTED: cationic amino acid transporter 7, chloroplastic-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_000176_050 |
0.085829219 |
- |
- |
glutamine amidotransferase class-I domain-containing family protein [Populus trichocarpa] |
| 9 |
Hb_002119_150 |
0.0867906908 |
- |
- |
hypothetical protein POPTR_0008s13870g [Populus trichocarpa] |
| 10 |
Hb_000402_060 |
0.0975698718 |
- |
- |
PREDICTED: uncharacterized protein LOC105644642 isoform X2 [Jatropha curcas] |
| 11 |
Hb_010883_160 |
0.1027121587 |
- |
- |
PREDICTED: pathogenesis-related protein 5 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000654_020 |
0.1066379353 |
- |
- |
hypothetical protein CICLE_v10006262mg [Citrus clementina] |
| 13 |
Hb_002400_300 |
0.1071030376 |
- |
- |
PREDICTED: synaptotagmin-4 [Jatropha curcas] |
| 14 |
Hb_006920_020 |
0.1081072402 |
- |
- |
endo-1,4-beta-glucanase, putative [Ricinus communis] |
| 15 |
Hb_001657_020 |
0.1081421929 |
- |
- |
heat shock family protein [Populus trichocarpa] |
| 16 |
Hb_000670_070 |
0.1084143031 |
- |
- |
PREDICTED: probable carbohydrate esterase At4g34215 [Jatropha curcas] |
| 17 |
Hb_008518_060 |
0.1103053522 |
- |
- |
PREDICTED: long chain acyl-CoA synthetase 1 [Jatropha curcas] |
| 18 |
Hb_001269_540 |
0.1113003171 |
- |
- |
hypothetical protein VITISV_038016 [Vitis vinifera] |
| 19 |
Hb_089124_010 |
0.1114157645 |
- |
- |
hypothetical protein POPTR_0018s09540g [Populus trichocarpa] |
| 20 |
Hb_003152_020 |
0.1126339349 |
- |
- |
PREDICTED: neutral alpha-glucosidase C [Jatropha curcas] |