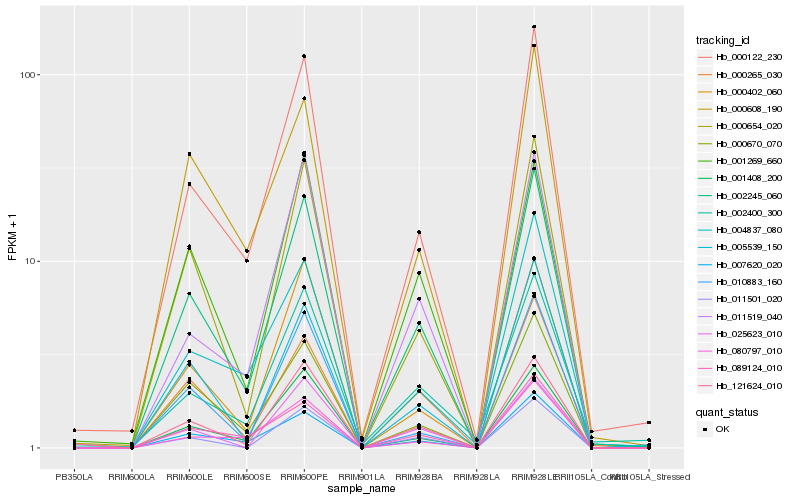
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002245_060 |
0.0 |
- |
- |
PREDICTED: kinesin-3 isoform X1 [Jatropha curcas] |
| 2 |
Hb_000122_230 |
0.0706094009 |
- |
- |
laccase, putative [Ricinus communis] |
| 3 |
Hb_000654_020 |
0.0768608035 |
- |
- |
hypothetical protein CICLE_v10006262mg [Citrus clementina] |
| 4 |
Hb_007620_020 |
0.0802877356 |
- |
- |
PREDICTED: cationic amino acid transporter 7, chloroplastic-like isoform X1 [Jatropha curcas] |
| 5 |
Hb_000670_070 |
0.0849363017 |
- |
- |
PREDICTED: probable carbohydrate esterase At4g34215 [Jatropha curcas] |
| 6 |
Hb_001408_200 |
0.087370141 |
- |
- |
hypothetical protein JCGZ_08964 [Jatropha curcas] |
| 7 |
Hb_005539_150 |
0.089679339 |
- |
- |
PREDICTED: probable xyloglucan endotransglucosylase/hydrolase protein 33 [Jatropha curcas] |
| 8 |
Hb_121624_010 |
0.0965288789 |
- |
- |
PREDICTED: uncharacterized protein LOC105634410 [Jatropha curcas] |
| 9 |
Hb_000265_030 |
0.0973245448 |
- |
- |
PREDICTED: subtilisin-like protease SBT5.3 [Jatropha curcas] |
| 10 |
Hb_000402_060 |
0.0995604893 |
- |
- |
PREDICTED: uncharacterized protein LOC105644642 isoform X2 [Jatropha curcas] |
| 11 |
Hb_011501_020 |
0.1017079911 |
- |
- |
PREDICTED: uncharacterized protein LOC105648929 [Jatropha curcas] |
| 12 |
Hb_089124_010 |
0.1040387721 |
- |
- |
hypothetical protein POPTR_0018s09540g [Populus trichocarpa] |
| 13 |
Hb_010883_160 |
0.1040470524 |
- |
- |
PREDICTED: pathogenesis-related protein 5 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000608_190 |
0.1086142004 |
rubber biosynthesis |
Gene Name: Isopentenyl-diphosphate delta isomerase |
PREDICTED: isopentenyl-diphosphate Delta-isomerase I [Jatropha curcas] |
| 15 |
Hb_001269_660 |
0.1138713622 |
- |
- |
PREDICTED: protein ASPARTIC PROTEASE IN GUARD CELL 2 [Jatropha curcas] |
| 16 |
Hb_011519_040 |
0.1160211073 |
- |
- |
hypothetical protein POPTR_0015s04770g [Populus trichocarpa] |
| 17 |
Hb_004837_080 |
0.1163232255 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB46 [Jatropha curcas] |
| 18 |
Hb_002400_300 |
0.1184396537 |
- |
- |
PREDICTED: synaptotagmin-4 [Jatropha curcas] |
| 19 |
Hb_080797_010 |
0.1200922811 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At3g47570-like [Citrus sinensis] |
| 20 |
Hb_025623_010 |
0.1232475641 |
- |
- |
- |