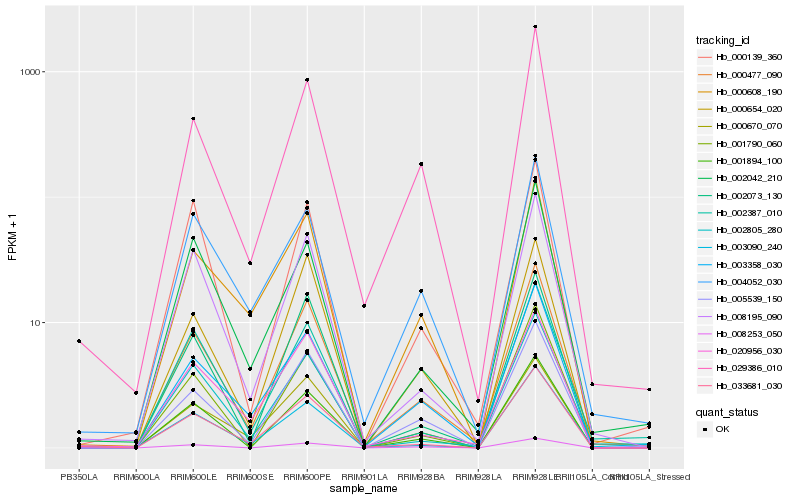
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000477_090 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105639302 [Jatropha curcas] |
| 2 |
Hb_005539_150 |
0.0730682431 |
- |
- |
PREDICTED: probable xyloglucan endotransglucosylase/hydrolase protein 33 [Jatropha curcas] |
| 3 |
Hb_008195_090 |
0.0839289149 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_001894_100 |
0.0889657294 |
- |
- |
PREDICTED: kininogen-1-like [Jatropha curcas] |
| 5 |
Hb_033681_030 |
0.0899013101 |
desease resistance |
Gene Name: NB-ARC |
Leucine-rich repeat containing protein [Theobroma cacao] |
| 6 |
Hb_029386_010 |
0.0899796132 |
- |
- |
RALF-LIKE 27 family protein [Populus trichocarpa] |
| 7 |
Hb_003358_030 |
0.0914790834 |
- |
- |
ankyrin-kinase, putative [Ricinus communis] |
| 8 |
Hb_002805_280 |
0.093380442 |
- |
- |
PREDICTED: sugar transport protein 14-like [Jatropha curcas] |
| 9 |
Hb_000670_070 |
0.0940022889 |
- |
- |
PREDICTED: probable carbohydrate esterase At4g34215 [Jatropha curcas] |
| 10 |
Hb_002073_130 |
0.094136656 |
- |
- |
PREDICTED: uncharacterized protein LOC100246830 [Vitis vinifera] |
| 11 |
Hb_008253_050 |
0.099548855 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0001s36220g [Populus trichocarpa] |
| 12 |
Hb_000654_020 |
0.1017805925 |
- |
- |
hypothetical protein CICLE_v10006262mg [Citrus clementina] |
| 13 |
Hb_000608_190 |
0.1055027832 |
rubber biosynthesis |
Gene Name: Isopentenyl-diphosphate delta isomerase |
PREDICTED: isopentenyl-diphosphate Delta-isomerase I [Jatropha curcas] |
| 14 |
Hb_001790_060 |
0.106428028 |
- |
- |
PREDICTED: uncharacterized protein LOC105644961 [Jatropha curcas] |
| 15 |
Hb_020956_030 |
0.1071851417 |
- |
- |
PREDICTED: swi5-dependent recombination DNA repair protein 1 homolog [Jatropha curcas] |
| 16 |
Hb_002387_010 |
0.1101518895 |
- |
- |
PREDICTED: protein STAY-GREEN LIKE, chloroplastic [Jatropha curcas] |
| 17 |
Hb_004052_030 |
0.1101543932 |
- |
- |
PREDICTED: uncharacterized protein LOC105640637 [Jatropha curcas] |
| 18 |
Hb_003090_240 |
0.1109533294 |
- |
- |
hypothetical protein JCGZ_25095 [Jatropha curcas] |
| 19 |
Hb_000139_360 |
0.1121003782 |
- |
- |
PREDICTED: uncharacterized protein LOC105631933 [Jatropha curcas] |
| 20 |
Hb_002042_210 |
0.1124573432 |
- |
- |
PREDICTED: UPF0603 protein At1g54780, chloroplastic [Jatropha curcas] |