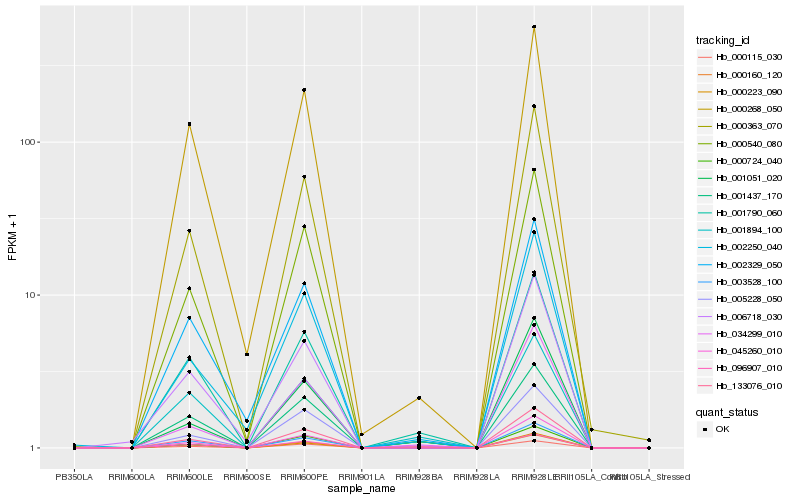
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_133076_010 |
0.0 |
- |
- |
PREDICTED: scopoletin glucosyltransferase-like [Populus euphratica] |
| 2 |
Hb_001790_060 |
0.0453275272 |
- |
- |
PREDICTED: uncharacterized protein LOC105644961 [Jatropha curcas] |
| 3 |
Hb_000540_080 |
0.0705973609 |
- |
- |
PREDICTED: laccase-17 [Jatropha curcas] |
| 4 |
Hb_000363_070 |
0.0832273252 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_001894_100 |
0.0838524748 |
- |
- |
PREDICTED: kininogen-1-like [Jatropha curcas] |
| 6 |
Hb_045260_010 |
0.0853705199 |
- |
- |
PREDICTED: leucine-rich repeat receptor protein kinase EMS1-like [Jatropha curcas] |
| 7 |
Hb_000268_050 |
0.0879508441 |
- |
- |
PREDICTED: tetrapyrrole-binding protein, chloroplastic [Jatropha curcas] |
| 8 |
Hb_000160_120 |
0.0905040118 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_096907_010 |
0.0926270474 |
- |
- |
PREDICTED: uncharacterized protein LOC104800023, partial [Tarenaya hassleriana] |
| 10 |
Hb_002329_050 |
0.0932892021 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000223_090 |
0.0936303464 |
- |
- |
Blue copper protein precursor, putative [Ricinus communis] |
| 12 |
Hb_001437_170 |
0.0950289892 |
- |
- |
hypothetical protein POPTR_0009s01390g [Populus trichocarpa] |
| 13 |
Hb_034299_010 |
0.0959379949 |
- |
- |
F-box and wd40 domain protein, putative [Ricinus communis] |
| 14 |
Hb_002250_040 |
0.0961420201 |
- |
- |
PREDICTED: transcription repressor OFP17 [Jatropha curcas] |
| 15 |
Hb_005228_050 |
0.0970313095 |
- |
- |
PREDICTED: uncharacterized protein LOC104896168 [Beta vulgaris subsp. vulgaris] |
| 16 |
Hb_006718_030 |
0.0981185845 |
- |
- |
ATPase, F1 complex, gamma subunit protein [Theobroma cacao] |
| 17 |
Hb_000115_030 |
0.0986253701 |
transcription factor |
TF Family: LOB |
PREDICTED: LOB domain-containing protein 42 [Jatropha curcas] |
| 18 |
Hb_000724_040 |
0.0987772193 |
transcription factor |
TF Family: MIKC |
PREDICTED: MADS-box protein CMB1-like [Jatropha curcas] |
| 19 |
Hb_001051_020 |
0.0990541145 |
- |
- |
hypothetical protein L484_008044 [Morus notabilis] |
| 20 |
Hb_003528_100 |
0.0996497574 |
- |
- |
beta-1,3-glucuronyltransferase, putative [Ricinus communis] |