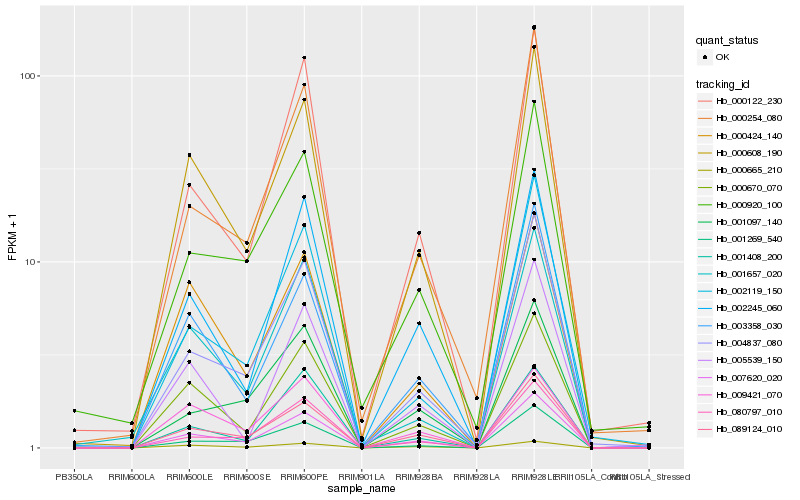
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007620_020 |
0.0 |
- |
- |
PREDICTED: cationic amino acid transporter 7, chloroplastic-like isoform X1 [Jatropha curcas] |
| 2 |
Hb_089124_010 |
0.039414038 |
- |
- |
hypothetical protein POPTR_0018s09540g [Populus trichocarpa] |
| 3 |
Hb_000608_190 |
0.0557580186 |
rubber biosynthesis |
Gene Name: Isopentenyl-diphosphate delta isomerase |
PREDICTED: isopentenyl-diphosphate Delta-isomerase I [Jatropha curcas] |
| 4 |
Hb_000670_070 |
0.0702531709 |
- |
- |
PREDICTED: probable carbohydrate esterase At4g34215 [Jatropha curcas] |
| 5 |
Hb_004837_080 |
0.0706854121 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB46 [Jatropha curcas] |
| 6 |
Hb_002245_060 |
0.0802877356 |
- |
- |
PREDICTED: kinesin-3 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000122_230 |
0.0841632256 |
- |
- |
laccase, putative [Ricinus communis] |
| 8 |
Hb_080797_010 |
0.0861228691 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At3g47570-like [Citrus sinensis] |
| 9 |
Hb_001269_540 |
0.0919491753 |
- |
- |
hypothetical protein VITISV_038016 [Vitis vinifera] |
| 10 |
Hb_003358_030 |
0.0942383426 |
- |
- |
ankyrin-kinase, putative [Ricinus communis] |
| 11 |
Hb_000254_080 |
0.0963151767 |
- |
- |
PREDICTED: uncharacterized protein LOC105635000 [Jatropha curcas] |
| 12 |
Hb_002119_150 |
0.0983903087 |
- |
- |
hypothetical protein POPTR_0008s13870g [Populus trichocarpa] |
| 13 |
Hb_000424_140 |
0.1045102627 |
- |
- |
Protein kinase APK1A, chloroplast precursor, putative [Ricinus communis] |
| 14 |
Hb_001657_020 |
0.1099487975 |
- |
- |
heat shock family protein [Populus trichocarpa] |
| 15 |
Hb_001097_140 |
0.1112259518 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000920_100 |
0.1132064162 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 17 |
Hb_000665_210 |
0.1148744492 |
- |
- |
PREDICTED: synaptotagmin-4 [Jatropha curcas] |
| 18 |
Hb_001408_200 |
0.1165031707 |
- |
- |
hypothetical protein JCGZ_08964 [Jatropha curcas] |
| 19 |
Hb_009421_070 |
0.1165507804 |
- |
- |
PREDICTED: cell number regulator 1-like [Jatropha curcas] |
| 20 |
Hb_005539_150 |
0.1168187764 |
- |
- |
PREDICTED: probable xyloglucan endotransglucosylase/hydrolase protein 33 [Jatropha curcas] |