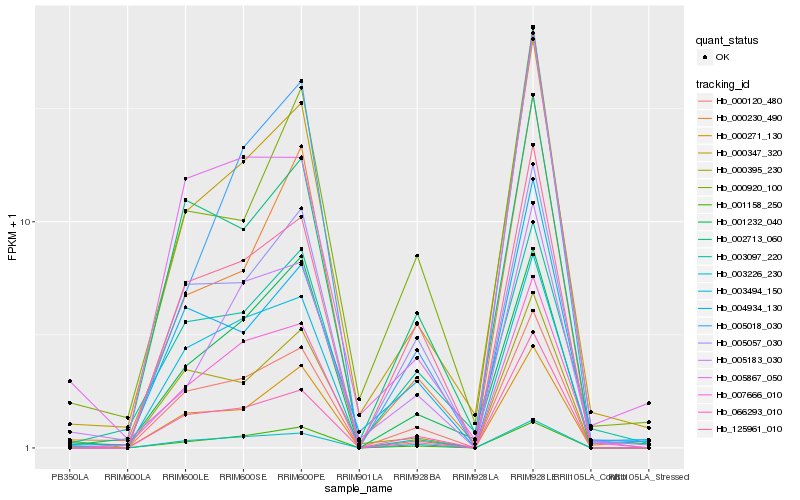
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000347_320 |
0.0 |
- |
- |
PREDICTED: subtilisin-like protease SBT1.7 [Jatropha curcas] |
| 2 |
Hb_000271_130 |
0.111387884 |
transcription factor |
TF Family: C2C2-Dof |
DNA binding with one finger 2.4, putative [Theobroma cacao] |
| 3 |
Hb_007666_010 |
0.1117026797 |
- |
- |
PREDICTED: uncharacterized protein LOC105649266 [Jatropha curcas] |
| 4 |
Hb_000120_480 |
0.1119579312 |
- |
- |
- |
| 5 |
Hb_000230_490 |
0.1122992665 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_066293_010 |
0.1156034989 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g35710 [Jatropha curcas] |
| 7 |
Hb_000920_100 |
0.1175878558 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 8 |
Hb_002713_060 |
0.118197993 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 6.4 [Jatropha curcas] |
| 9 |
Hb_005183_030 |
0.1193391492 |
- |
- |
PREDICTED: probable calcium-binding protein CML16 [Jatropha curcas] |
| 10 |
Hb_004934_130 |
0.1201451401 |
- |
- |
12-oxophytodienoate reductase 2 isoform 1 [Theobroma cacao] |
| 11 |
Hb_005057_030 |
0.1210982816 |
- |
- |
Leucoanthocyanidin dioxygenase, putative [Ricinus communis] |
| 12 |
Hb_001232_040 |
0.124771631 |
- |
- |
PREDICTED: uncharacterized protein LOC105634974 [Jatropha curcas] |
| 13 |
Hb_001158_250 |
0.1252008958 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_003494_150 |
0.1252350981 |
- |
- |
Silicon transporter, putative [Ricinus communis] |
| 15 |
Hb_000395_230 |
0.1267981849 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase 1 [Vitis vinifera] |
| 16 |
Hb_003226_230 |
0.1268029542 |
- |
- |
PREDICTED: transposon Ty3-G Gag-Pol polyprotein [Phoenix dactylifera] |
| 17 |
Hb_003097_220 |
0.1279015332 |
- |
- |
hypothetical protein POPTR_0001s06150g [Populus trichocarpa] |
| 18 |
Hb_125961_010 |
0.1312469972 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 19 |
Hb_005018_030 |
0.1320990971 |
- |
- |
hypothetical protein CICLE_v10019811mg [Citrus clementina] |
| 20 |
Hb_005867_050 |
0.1324000316 |
- |
- |
PREDICTED: protein ECERIFERUM 1 [Jatropha curcas] |