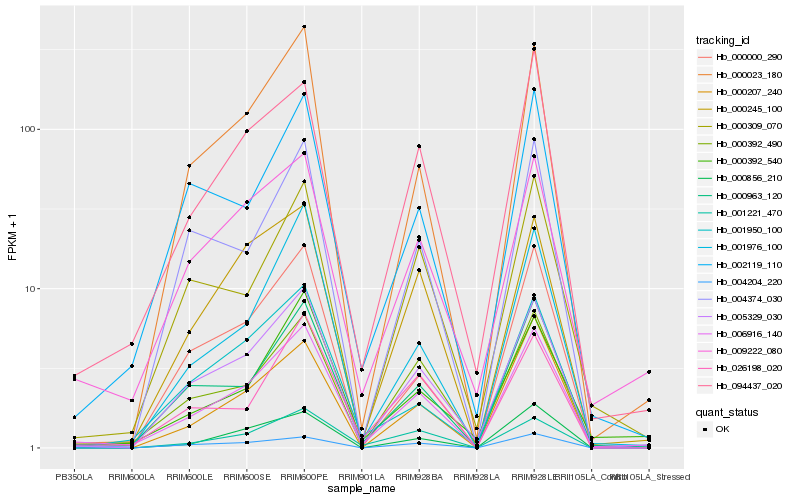
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006916_140 |
0.0 |
- |
- |
PREDICTED: PI-PLC X domain-containing protein At5g67130-like [Jatropha curcas] |
| 2 |
Hb_000963_120 |
0.0953016841 |
- |
- |
PREDICTED: uncharacterized protein LOC105628103 isoform X1 [Jatropha curcas] |
| 3 |
Hb_005329_030 |
0.1011629921 |
- |
- |
PREDICTED: NAD(P)H dehydrogenase (quinone) FQR1-like [Jatropha curcas] |
| 4 |
Hb_009222_080 |
0.105846068 |
- |
- |
RING-H2 finger protein ATL2J, putative [Ricinus communis] |
| 5 |
Hb_000392_490 |
0.1084715978 |
- |
- |
PREDICTED: uncharacterized protein LOC105640495 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000023_180 |
0.1110736743 |
- |
- |
Aspartic proteinase nepenthesin-2 precursor, putative [Ricinus communis] |
| 7 |
Hb_094437_020 |
0.1146862948 |
- |
- |
Xylem serine proteinase 1 precursor, putative [Ricinus communis] |
| 8 |
Hb_000245_100 |
0.1275971257 |
- |
- |
PREDICTED: uncharacterized protein LOC105631555 [Jatropha curcas] |
| 9 |
Hb_002119_110 |
0.1318442055 |
- |
- |
PREDICTED: probable xyloglucan endotransglucosylase/hydrolase protein 28 [Jatropha curcas] |
| 10 |
Hb_000000_290 |
0.1322947597 |
- |
- |
PREDICTED: probable 1-acyl-sn-glycerol-3-phosphate acyltransferase 4 [Jatropha curcas] |
| 11 |
Hb_000392_540 |
0.1334387446 |
- |
- |
PREDICTED: hypersensitive-induced response protein 1 [Sesamum indicum] |
| 12 |
Hb_000856_210 |
0.1338774085 |
- |
- |
nitrate transporter, putative [Ricinus communis] |
| 13 |
Hb_000309_070 |
0.1339405662 |
- |
- |
PREDICTED: 2-methylene-furan-3-one reductase [Cucumis melo] |
| 14 |
Hb_001976_100 |
0.1348208401 |
- |
- |
PREDICTED: receptor-like cytosolic serine/threonine-protein kinase RBK1 isoform X2 [Jatropha curcas] |
| 15 |
Hb_026198_020 |
0.1350003703 |
- |
- |
PREDICTED: uncharacterized protein LOC105634368 [Jatropha curcas] |
| 16 |
Hb_000207_240 |
0.1399596329 |
- |
- |
hypothetical protein RCOM_0686540 [Ricinus communis] |
| 17 |
Hb_001221_470 |
0.1401975689 |
- |
- |
PREDICTED: uncharacterized protein LOC105648774 [Jatropha curcas] |
| 18 |
Hb_001950_100 |
0.1404349387 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g37250 [Jatropha curcas] |
| 19 |
Hb_004374_030 |
0.1406694535 |
- |
- |
PREDICTED: probable polygalacturonase [Jatropha curcas] |
| 20 |
Hb_004204_220 |
0.1412437169 |
- |
- |
- |