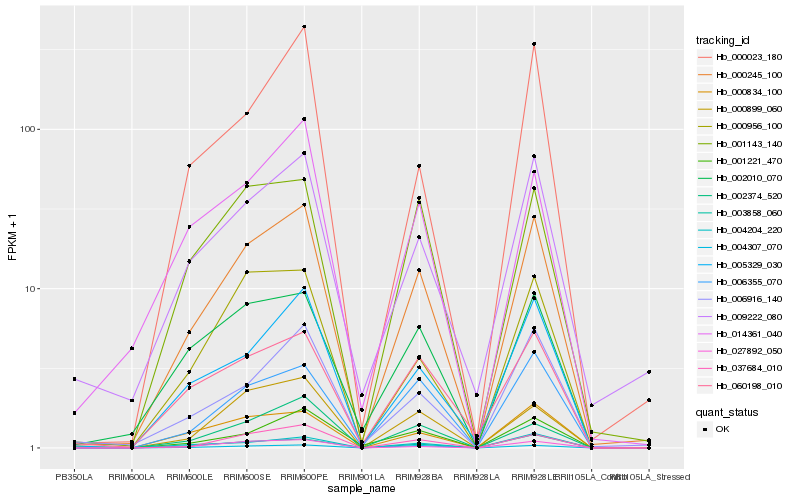
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000245_100 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105631555 [Jatropha curcas] |
| 2 |
Hb_005329_030 |
0.1168852098 |
- |
- |
PREDICTED: NAD(P)H dehydrogenase (quinone) FQR1-like [Jatropha curcas] |
| 3 |
Hb_037684_010 |
0.1184332516 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105645015 [Jatropha curcas] |
| 4 |
Hb_003858_060 |
0.1189827812 |
- |
- |
Germin-like protein subfamily 1 member 18 [Theobroma cacao] |
| 5 |
Hb_006355_070 |
0.1197988101 |
transcription factor |
TF Family: bHLH |
Phytochrome-interacting factor, putative [Ricinus communis] |
| 6 |
Hb_004204_220 |
0.1254939393 |
- |
- |
- |
| 7 |
Hb_009222_080 |
0.1268707866 |
- |
- |
RING-H2 finger protein ATL2J, putative [Ricinus communis] |
| 8 |
Hb_004307_070 |
0.1273514601 |
- |
- |
kinase, putative [Ricinus communis] |
| 9 |
Hb_006916_140 |
0.1275971257 |
- |
- |
PREDICTED: PI-PLC X domain-containing protein At5g67130-like [Jatropha curcas] |
| 10 |
Hb_000834_100 |
0.1301597012 |
- |
- |
PREDICTED: uncharacterized protein LOC105178355 [Sesamum indicum] |
| 11 |
Hb_000956_100 |
0.1312329731 |
- |
- |
Cucumisin precursor, putative [Ricinus communis] |
| 12 |
Hb_001221_470 |
0.1312946162 |
- |
- |
PREDICTED: uncharacterized protein LOC105648774 [Jatropha curcas] |
| 13 |
Hb_001143_140 |
0.1330690166 |
- |
- |
Aldehyde dehydrogenase, putative [Ricinus communis] |
| 14 |
Hb_027892_050 |
0.133203198 |
- |
- |
hypothetical protein PHAVU_009G226400g [Phaseolus vulgaris] |
| 15 |
Hb_060198_010 |
0.1372890932 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RGA2-like [Jatropha curcas] |
| 16 |
Hb_002010_070 |
0.1373805285 |
- |
- |
hypothetical protein EUGRSUZ_I027342, partial [Eucalyptus grandis] |
| 17 |
Hb_000899_060 |
0.1375054166 |
- |
- |
PREDICTED: uncharacterized protein LOC105793631 [Gossypium raimondii] |
| 18 |
Hb_002374_520 |
0.1377178451 |
- |
- |
early nodulin 8 precursor family protein [Populus trichocarpa] |
| 19 |
Hb_014361_040 |
0.1404376478 |
- |
- |
PREDICTED: subtilisin-like protease SBT1.7 [Jatropha curcas] |
| 20 |
Hb_000023_180 |
0.1448960065 |
- |
- |
Aspartic proteinase nepenthesin-2 precursor, putative [Ricinus communis] |