| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
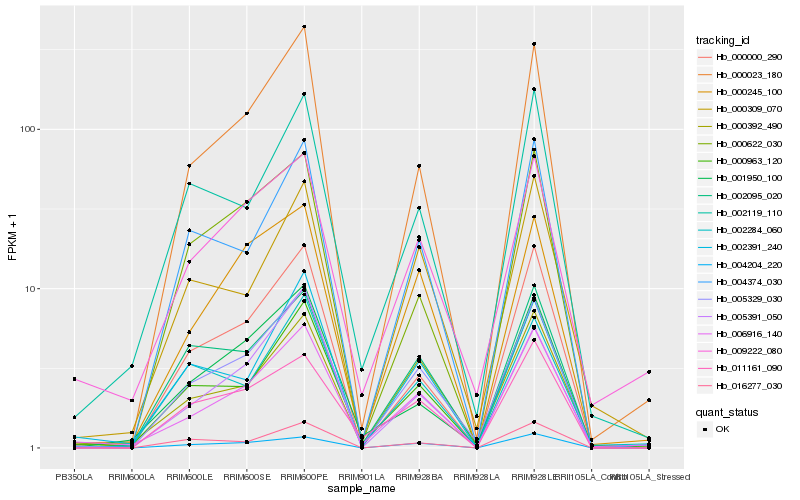
Hb_005329_030 |
0.0 |
- |
- |
PREDICTED: NAD(P)H dehydrogenase (quinone) FQR1-like [Jatropha curcas] |
| 2 |
Hb_000023_180 |
0.0849181435 |
- |
- |
Aspartic proteinase nepenthesin-2 precursor, putative [Ricinus communis] |
| 3 |
Hb_006916_140 |
0.1011629921 |
- |
- |
PREDICTED: PI-PLC X domain-containing protein At5g67130-like [Jatropha curcas] |
| 4 |
Hb_004374_030 |
0.1062464887 |
- |
- |
PREDICTED: probable polygalacturonase [Jatropha curcas] |
| 5 |
Hb_000963_120 |
0.1104063308 |
- |
- |
PREDICTED: uncharacterized protein LOC105628103 isoform X1 [Jatropha curcas] |
| 6 |
Hb_004204_220 |
0.1109002176 |
- |
- |
- |
| 7 |
Hb_000000_290 |
0.1111250111 |
- |
- |
PREDICTED: probable 1-acyl-sn-glycerol-3-phosphate acyltransferase 4 [Jatropha curcas] |
| 8 |
Hb_000392_490 |
0.1113576168 |
- |
- |
PREDICTED: uncharacterized protein LOC105640495 isoform X1 [Jatropha curcas] |
| 9 |
Hb_002095_020 |
0.112936024 |
- |
- |
PREDICTED: glutamate receptor 3.4-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_000245_100 |
0.1168852098 |
- |
- |
PREDICTED: uncharacterized protein LOC105631555 [Jatropha curcas] |
| 11 |
Hb_005391_050 |
0.1205572339 |
- |
- |
PREDICTED: uncharacterized protein LOC105643654 [Jatropha curcas] |
| 12 |
Hb_016277_030 |
0.1218085136 |
- |
- |
hypothetical protein JCGZ_16973 [Jatropha curcas] |
| 13 |
Hb_009222_080 |
0.1259021915 |
- |
- |
RING-H2 finger protein ATL2J, putative [Ricinus communis] |
| 14 |
Hb_001950_100 |
0.1260173987 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g37250 [Jatropha curcas] |
| 15 |
Hb_002391_240 |
0.1262929843 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_002119_110 |
0.1264831317 |
- |
- |
PREDICTED: probable xyloglucan endotransglucosylase/hydrolase protein 28 [Jatropha curcas] |
| 17 |
Hb_000622_030 |
0.1267996387 |
- |
- |
PREDICTED: hydroquinone glucosyltransferase [Jatropha curcas] |
| 18 |
Hb_011161_090 |
0.1272628723 |
- |
- |
- |
| 19 |
Hb_000309_070 |
0.1303668038 |
- |
- |
PREDICTED: 2-methylene-furan-3-one reductase [Cucumis melo] |
| 20 |
Hb_002284_060 |
0.1314544009 |
- |
- |
sugar transporter, putative [Ricinus communis] |