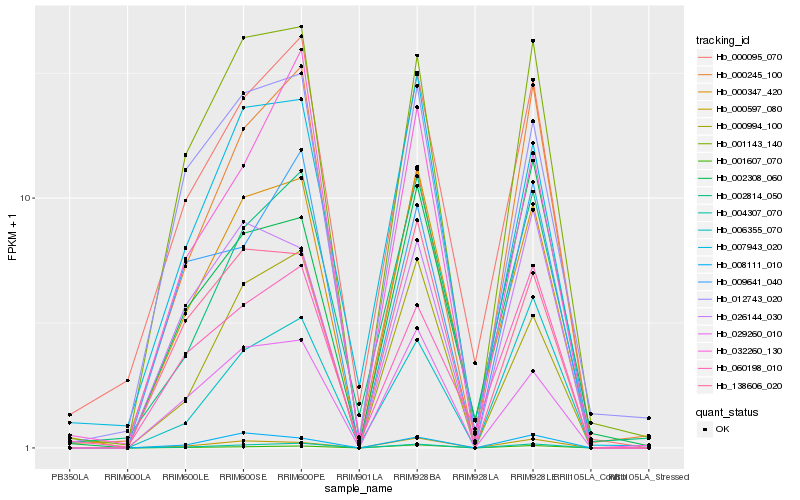
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004307_070 |
0.0 |
- |
- |
kinase, putative [Ricinus communis] |
| 2 |
Hb_001143_140 |
0.0864824607 |
- |
- |
Aldehyde dehydrogenase, putative [Ricinus communis] |
| 3 |
Hb_000347_420 |
0.1001514028 |
- |
- |
hypothetical protein JCGZ_24696 [Jatropha curcas] |
| 4 |
Hb_006355_070 |
0.1058075933 |
transcription factor |
TF Family: bHLH |
Phytochrome-interacting factor, putative [Ricinus communis] |
| 5 |
Hb_000095_070 |
0.1136218128 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_009641_040 |
0.116379028 |
- |
- |
PREDICTED: uncharacterized protein LOC105633355 [Jatropha curcas] |
| 7 |
Hb_000994_100 |
0.119539179 |
- |
- |
RALFL33, putative [Ricinus communis] |
| 8 |
Hb_002814_050 |
0.1222618608 |
- |
- |
PREDICTED: sugar carrier protein A [Jatropha curcas] |
| 9 |
Hb_000245_100 |
0.1273514601 |
- |
- |
PREDICTED: uncharacterized protein LOC105631555 [Jatropha curcas] |
| 10 |
Hb_029260_010 |
0.1290345416 |
- |
- |
nitrate transporter, putative [Ricinus communis] |
| 11 |
Hb_060198_010 |
0.1309275112 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RGA2-like [Jatropha curcas] |
| 12 |
Hb_001607_070 |
0.1309598767 |
- |
- |
hypothetical protein JCGZ_09913 [Jatropha curcas] |
| 13 |
Hb_000597_080 |
0.1336923169 |
- |
- |
PREDICTED: 7-deoxyloganetic acid glucosyltransferase-like isoform X2 [Jatropha curcas] |
| 14 |
Hb_026144_030 |
0.1344180888 |
- |
- |
PREDICTED: high affinity nitrate transporter 2.5 [Jatropha curcas] |
| 15 |
Hb_008111_010 |
0.1345749845 |
- |
- |
hypothetical protein VITISV_021527 [Vitis vinifera] |
| 16 |
Hb_012743_020 |
0.1365183278 |
- |
- |
serine/threonine protein phosphatase, putative [Ricinus communis] |
| 17 |
Hb_138606_020 |
0.1386833067 |
- |
- |
Nitrate transporter2.5 [Theobroma cacao] |
| 18 |
Hb_032260_130 |
0.1424104127 |
- |
- |
hypothetical protein POPTR_0003s13800g [Populus trichocarpa] |
| 19 |
Hb_002308_060 |
0.1449516581 |
- |
- |
PREDICTED: cytochrome P450 704C1-like [Jatropha curcas] |
| 20 |
Hb_007943_020 |
0.1451599985 |
- |
- |
transcription factor, putative [Ricinus communis] |