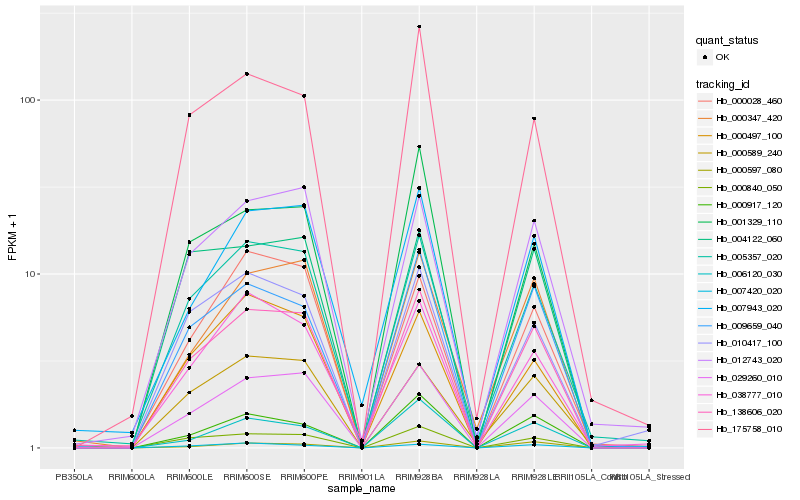
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_138606_020 |
0.0 |
- |
- |
Nitrate transporter2.5 [Theobroma cacao] |
| 2 |
Hb_029260_010 |
0.043313735 |
- |
- |
nitrate transporter, putative [Ricinus communis] |
| 3 |
Hb_000840_050 |
0.0731322663 |
desease resistance |
Gene Name: TIR |
PREDICTED: TMV resistance protein N-like isoform X2 [Vitis vinifera] |
| 4 |
Hb_009659_040 |
0.085868984 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein NUTCRACKER [Populus euphratica] |
| 5 |
Hb_005357_020 |
0.0885667039 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000347_420 |
0.1020659752 |
- |
- |
hypothetical protein JCGZ_24696 [Jatropha curcas] |
| 7 |
Hb_012743_020 |
0.1022457836 |
- |
- |
serine/threonine protein phosphatase, putative [Ricinus communis] |
| 8 |
Hb_000917_120 |
0.1086026708 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_007943_020 |
0.1111959255 |
- |
- |
transcription factor, putative [Ricinus communis] |
| 10 |
Hb_004122_060 |
0.1131526938 |
- |
- |
PREDICTED: uncharacterized acetyltransferase At3g50280-like [Jatropha curcas] |
| 11 |
Hb_007420_020 |
0.1132735246 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 12 |
Hb_010417_100 |
0.1147668698 |
- |
- |
PREDICTED: respiratory burst oxidase homolog protein A [Jatropha curcas] |
| 13 |
Hb_000589_240 |
0.1169256323 |
- |
- |
Aspartic proteinase nepenthesin-1 precursor, putative [Ricinus communis] |
| 14 |
Hb_175758_010 |
0.1177241162 |
- |
- |
PREDICTED: aspartic proteinase A1-like isoform X2 [Jatropha curcas] |
| 15 |
Hb_001329_110 |
0.1209079482 |
- |
- |
PREDICTED: uncharacterized protein LOC105642675 [Jatropha curcas] |
| 16 |
Hb_000597_080 |
0.1211775429 |
- |
- |
PREDICTED: 7-deoxyloganetic acid glucosyltransferase-like isoform X2 [Jatropha curcas] |
| 17 |
Hb_006120_030 |
0.122211138 |
- |
- |
- |
| 18 |
Hb_000497_100 |
0.1224125015 |
- |
- |
respiratory burst oxidase D [Manihot esculenta] |
| 19 |
Hb_000028_460 |
0.1232580974 |
- |
- |
hypothetical protein POPTR_0015s05340g [Populus trichocarpa] |
| 20 |
Hb_038777_010 |
0.1238582029 |
transcription factor |
TF Family: G2-like |
PREDICTED: probable transcription factor KAN2 isoform X1 [Populus euphratica] |