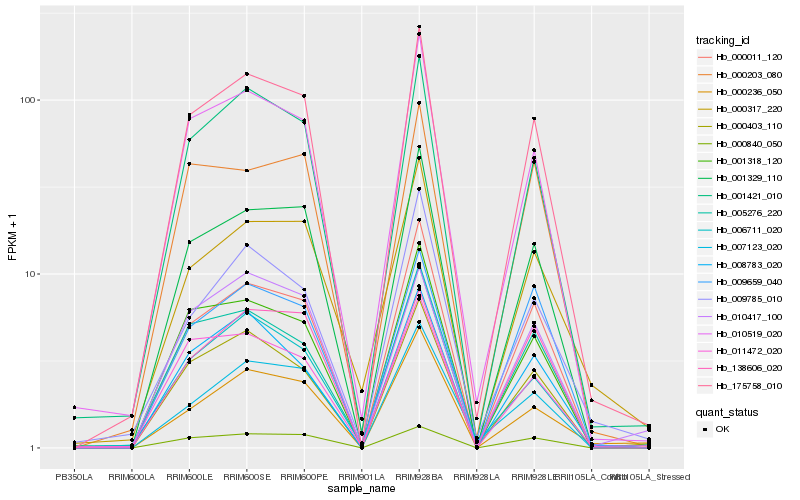
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_175758_010 |
0.0 |
- |
- |
PREDICTED: aspartic proteinase A1-like isoform X2 [Jatropha curcas] |
| 2 |
Hb_001421_010 |
0.0528134672 |
transcription factor |
TF Family: ARR-B |
PREDICTED: two-component response regulator-like APRR2 isoform X1 [Jatropha curcas] |
| 3 |
Hb_001329_110 |
0.0602989902 |
- |
- |
PREDICTED: uncharacterized protein LOC105642675 [Jatropha curcas] |
| 4 |
Hb_010519_020 |
0.0649726801 |
- |
- |
hypothetical protein JCGZ_19028 [Jatropha curcas] |
| 5 |
Hb_000011_120 |
0.074233643 |
- |
- |
PREDICTED: ATPase 10, plasma membrane-type-like [Populus euphratica] |
| 6 |
Hb_001318_120 |
0.0757748559 |
- |
- |
PREDICTED: uncharacterized protein LOC105642675 [Jatropha curcas] |
| 7 |
Hb_000840_050 |
0.08084866 |
desease resistance |
Gene Name: TIR |
PREDICTED: TMV resistance protein N-like isoform X2 [Vitis vinifera] |
| 8 |
Hb_000403_110 |
0.0808594559 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 51 [Jatropha curcas] |
| 9 |
Hb_005276_220 |
0.0852632725 |
transcription factor |
TF Family: bHLH |
PREDICTED: basic helix-loop-helix protein A [Jatropha curcas] |
| 10 |
Hb_011472_020 |
0.09098758 |
- |
- |
PREDICTED: mucin-17-like [Jatropha curcas] |
| 11 |
Hb_008783_020 |
0.0922212083 |
- |
- |
hypothetical protein JCGZ_21284 [Jatropha curcas] |
| 12 |
Hb_007123_020 |
0.0985522553 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein HOX3 [Jatropha curcas] |
| 13 |
Hb_000203_080 |
0.104218652 |
- |
- |
Xylem bark cysteine peptidase 3 isoform 1 [Theobroma cacao] |
| 14 |
Hb_009659_040 |
0.1079235809 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein NUTCRACKER [Populus euphratica] |
| 15 |
Hb_006711_020 |
0.1098282493 |
- |
- |
hypothetical protein JCGZ_07258 [Jatropha curcas] |
| 16 |
Hb_000236_050 |
0.1134993309 |
- |
- |
phosphate transporter [Manihot esculenta] |
| 17 |
Hb_010417_100 |
0.1143671598 |
- |
- |
PREDICTED: respiratory burst oxidase homolog protein A [Jatropha curcas] |
| 18 |
Hb_000317_220 |
0.1158533414 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_009785_010 |
0.1172169653 |
- |
- |
PREDICTED: uncharacterized protein LOC105648749 [Jatropha curcas] |
| 20 |
Hb_138606_020 |
0.1177241162 |
- |
- |
Nitrate transporter2.5 [Theobroma cacao] |