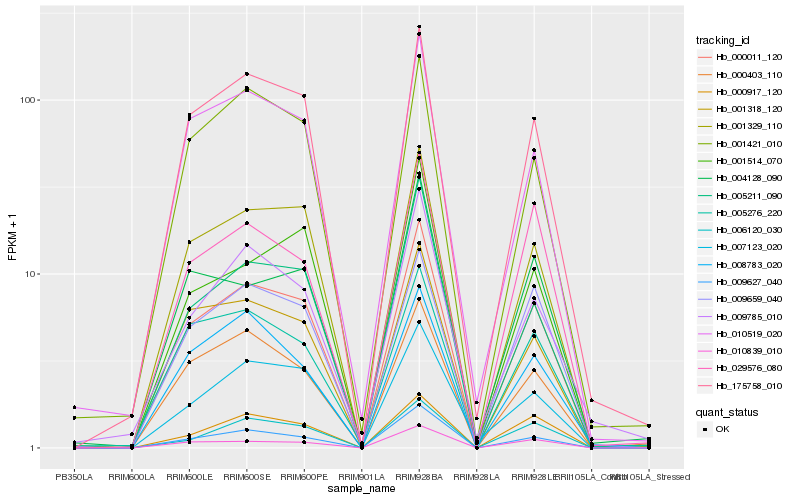
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000011_120 |
0.0 |
- |
- |
PREDICTED: ATPase 10, plasma membrane-type-like [Populus euphratica] |
| 2 |
Hb_001329_110 |
0.0656768334 |
- |
- |
PREDICTED: uncharacterized protein LOC105642675 [Jatropha curcas] |
| 3 |
Hb_009627_040 |
0.0734351414 |
- |
- |
Lipase class 3-related protein, putative [Theobroma cacao] |
| 4 |
Hb_175758_010 |
0.074233643 |
- |
- |
PREDICTED: aspartic proteinase A1-like isoform X2 [Jatropha curcas] |
| 5 |
Hb_005211_090 |
0.0831608681 |
- |
- |
PREDICTED: aluminum-activated malate transporter 4-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_010839_010 |
0.0868906057 |
- |
- |
receptor-kinase, putative [Ricinus communis] |
| 7 |
Hb_001318_120 |
0.0882029029 |
- |
- |
PREDICTED: uncharacterized protein LOC105642675 [Jatropha curcas] |
| 8 |
Hb_010519_020 |
0.0937352352 |
- |
- |
hypothetical protein JCGZ_19028 [Jatropha curcas] |
| 9 |
Hb_000917_120 |
0.0960487113 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_006120_030 |
0.1005427472 |
- |
- |
- |
| 11 |
Hb_000403_110 |
0.100635087 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 51 [Jatropha curcas] |
| 12 |
Hb_007123_020 |
0.1009799405 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein HOX3 [Jatropha curcas] |
| 13 |
Hb_029576_080 |
0.1045369251 |
- |
- |
Aspartic proteinase precursor, putative [Ricinus communis] |
| 14 |
Hb_001421_010 |
0.1071167571 |
transcription factor |
TF Family: ARR-B |
PREDICTED: two-component response regulator-like APRR2 isoform X1 [Jatropha curcas] |
| 15 |
Hb_005276_220 |
0.1072934516 |
transcription factor |
TF Family: bHLH |
PREDICTED: basic helix-loop-helix protein A [Jatropha curcas] |
| 16 |
Hb_001514_070 |
0.1093594071 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 17 |
Hb_009659_040 |
0.1109439385 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein NUTCRACKER [Populus euphratica] |
| 18 |
Hb_009785_010 |
0.1136571591 |
- |
- |
PREDICTED: uncharacterized protein LOC105648749 [Jatropha curcas] |
| 19 |
Hb_008783_020 |
0.1159106525 |
- |
- |
hypothetical protein JCGZ_21284 [Jatropha curcas] |
| 20 |
Hb_004128_090 |
0.1160848198 |
transcription factor |
TF Family: WRKY |
PREDICTED: WRKY transcription factor 44 [Jatropha curcas] |