| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
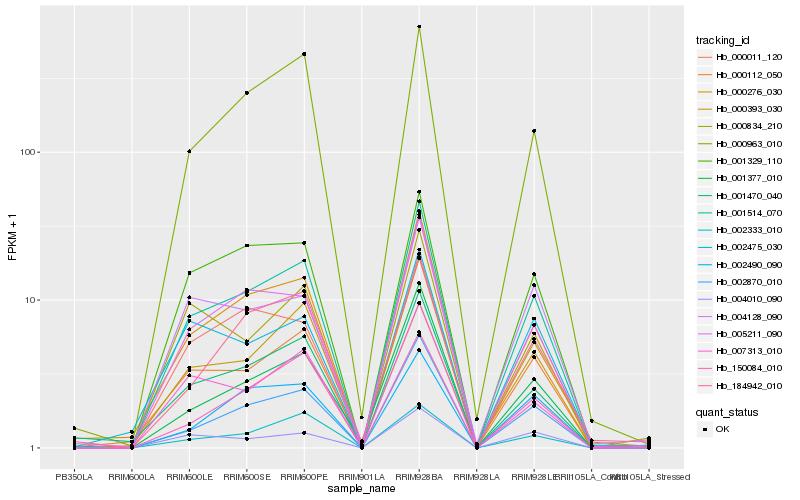
Hb_001514_070 |
0.0 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 2 |
Hb_000393_030 |
0.0761761147 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 3 |
Hb_002870_010 |
0.0820821337 |
- |
- |
PREDICTED: uncharacterized protein LOC105649339 [Jatropha curcas] |
| 4 |
Hb_005211_090 |
0.0829463166 |
- |
- |
PREDICTED: aluminum-activated malate transporter 4-like isoform X1 [Jatropha curcas] |
| 5 |
Hb_001470_040 |
0.0991340093 |
- |
- |
PREDICTED: probable BOI-related E3 ubiquitin-protein ligase 3 [Jatropha curcas] |
| 6 |
Hb_001329_110 |
0.1029777092 |
- |
- |
PREDICTED: uncharacterized protein LOC105642675 [Jatropha curcas] |
| 7 |
Hb_000963_010 |
0.1075482795 |
- |
- |
PREDICTED: aquaporin PIP2-4 [Jatropha curcas] |
| 8 |
Hb_007313_010 |
0.1081675145 |
- |
- |
hypothetical protein JCGZ_23398 [Jatropha curcas] |
| 9 |
Hb_001377_010 |
0.1091544478 |
- |
- |
PREDICTED: transcription factor TRY-like [Jatropha curcas] |
| 10 |
Hb_000011_120 |
0.1093594071 |
- |
- |
PREDICTED: ATPase 10, plasma membrane-type-like [Populus euphratica] |
| 11 |
Hb_000276_030 |
0.1106808259 |
- |
- |
dimethylaniline monooxygenase, putative [Ricinus communis] |
| 12 |
Hb_004010_090 |
0.1131185605 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 13 |
Hb_150084_010 |
0.1190746556 |
- |
- |
PREDICTED: premnaspirodiene oxygenase-like [Vitis vinifera] |
| 14 |
Hb_000834_210 |
0.1193434773 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 15 |
Hb_004128_090 |
0.1199942177 |
transcription factor |
TF Family: WRKY |
PREDICTED: WRKY transcription factor 44 [Jatropha curcas] |
| 16 |
Hb_000112_050 |
0.120618601 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor ASIL1 [Jatropha curcas] |
| 17 |
Hb_002333_010 |
0.1227156306 |
- |
- |
hypothetical protein JCGZ_18834 [Jatropha curcas] |
| 18 |
Hb_002490_090 |
0.1254209146 |
- |
- |
PREDICTED: uncharacterized protein LOC105133003 [Populus euphratica] |
| 19 |
Hb_184942_010 |
0.1277453093 |
- |
- |
STS14 protein precursor, putative [Ricinus communis] |
| 20 |
Hb_002475_030 |
0.1280105139 |
- |
- |
conserved hypothetical protein [Ricinus communis] |