| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
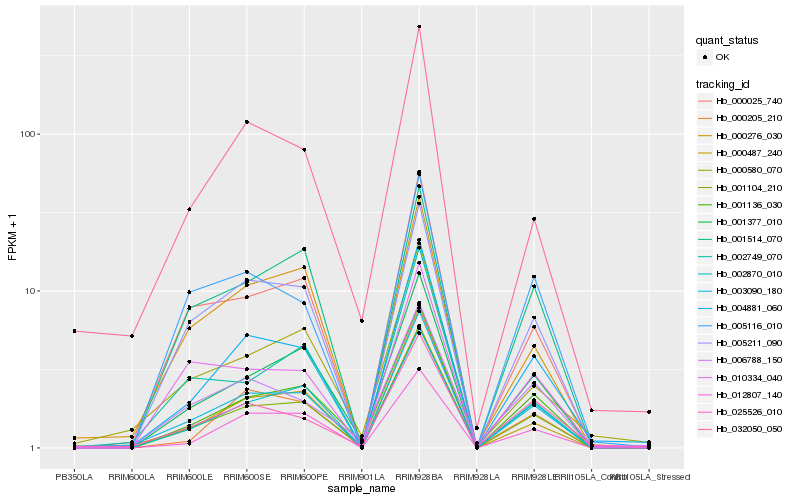
Hb_003090_180 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105646674 [Jatropha curcas] |
| 2 |
Hb_001104_210 |
0.0649604916 |
- |
- |
hypothetical protein JCGZ_23620 [Jatropha curcas] |
| 3 |
Hb_004881_060 |
0.0712524234 |
transcription factor |
TF Family: WRKY |
hypothetical protein POPTR_0005s22480g [Populus trichocarpa] |
| 4 |
Hb_000487_240 |
0.0833648196 |
- |
- |
PREDICTED: CYSTM1 family protein B [Cucumis melo] |
| 5 |
Hb_000025_740 |
0.0967818973 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12 isoform X1 [Jatropha curcas] |
| 6 |
Hb_001136_030 |
0.1016446925 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 25-like [Jatropha curcas] |
| 7 |
Hb_002870_010 |
0.1025098077 |
- |
- |
PREDICTED: uncharacterized protein LOC105649339 [Jatropha curcas] |
| 8 |
Hb_001377_010 |
0.1092995203 |
- |
- |
PREDICTED: transcription factor TRY-like [Jatropha curcas] |
| 9 |
Hb_005211_090 |
0.1125097602 |
- |
- |
PREDICTED: aluminum-activated malate transporter 4-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_000276_030 |
0.1166043585 |
- |
- |
dimethylaniline monooxygenase, putative [Ricinus communis] |
| 11 |
Hb_000580_070 |
0.1190085172 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At5g10290 [Jatropha curcas] |
| 12 |
Hb_032050_050 |
0.1206547104 |
- |
- |
PREDICTED: peroxidase 12-like [Jatropha curcas] |
| 13 |
Hb_025526_010 |
0.1216047003 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104452086 [Eucalyptus grandis] |
| 14 |
Hb_000205_210 |
0.1272815752 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein At4g27190-like isoform X1 [Citrus sinensis] |
| 15 |
Hb_005116_010 |
0.1277023762 |
transcription factor |
TF Family: bHLH |
PREDICTED: basic helix-loop-helix protein A [Jatropha curcas] |
| 16 |
Hb_010334_040 |
0.1289079247 |
- |
- |
PREDICTED: uncharacterized protein LOC105108579 [Populus euphratica] |
| 17 |
Hb_012807_140 |
0.1296609627 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0009s01270g [Populus trichocarpa] |
| 18 |
Hb_006788_150 |
0.1321231317 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_002749_070 |
0.1323595641 |
- |
- |
PREDICTED: uncharacterized protein LOC105635941 [Jatropha curcas] |
| 20 |
Hb_001514_070 |
0.1324710515 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |