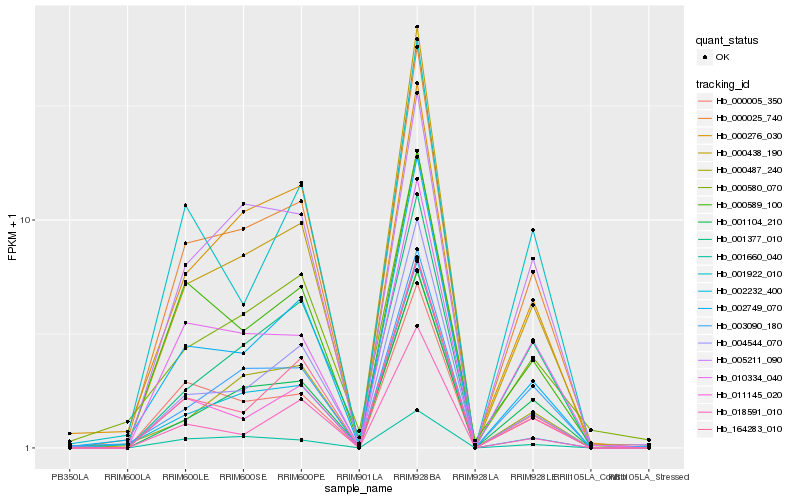
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000025_740 |
0.0 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12 isoform X1 [Jatropha curcas] |
| 2 |
Hb_002749_070 |
0.0776267862 |
- |
- |
PREDICTED: uncharacterized protein LOC105635941 [Jatropha curcas] |
| 3 |
Hb_010334_040 |
0.083841567 |
- |
- |
PREDICTED: uncharacterized protein LOC105108579 [Populus euphratica] |
| 4 |
Hb_000005_350 |
0.0872548572 |
- |
- |
PREDICTED: uncharacterized protein LOC105634263 [Jatropha curcas] |
| 5 |
Hb_001104_210 |
0.0907647588 |
- |
- |
hypothetical protein JCGZ_23620 [Jatropha curcas] |
| 6 |
Hb_004544_070 |
0.0961382567 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 7 |
Hb_164283_010 |
0.0961586562 |
- |
- |
hypothetical protein RCOM_0725500 [Ricinus communis] |
| 8 |
Hb_003090_180 |
0.0967818973 |
- |
- |
PREDICTED: uncharacterized protein LOC105646674 [Jatropha curcas] |
| 9 |
Hb_000589_100 |
0.0995939231 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 10 |
Hb_011145_020 |
0.1010244756 |
- |
- |
PREDICTED: metalloendoproteinase 1-like [Pyrus x bretschneideri] |
| 11 |
Hb_000487_240 |
0.1018781143 |
- |
- |
PREDICTED: CYSTM1 family protein B [Cucumis melo] |
| 12 |
Hb_001660_040 |
0.1091481683 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g08850 [Jatropha curcas] |
| 13 |
Hb_000438_190 |
0.1095631399 |
- |
- |
PREDICTED: peroxidase 4-like [Jatropha curcas] |
| 14 |
Hb_000276_030 |
0.1126201474 |
- |
- |
dimethylaniline monooxygenase, putative [Ricinus communis] |
| 15 |
Hb_000580_070 |
0.1159961391 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At5g10290 [Jatropha curcas] |
| 16 |
Hb_005211_090 |
0.1209782722 |
- |
- |
PREDICTED: aluminum-activated malate transporter 4-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_001377_010 |
0.1210999197 |
- |
- |
PREDICTED: transcription factor TRY-like [Jatropha curcas] |
| 18 |
Hb_001922_010 |
0.122636257 |
- |
- |
PREDICTED: ABC transporter G family member 8 [Jatropha curcas] |
| 19 |
Hb_002232_400 |
0.1227677114 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH25-like [Jatropha curcas] |
| 20 |
Hb_018591_010 |
0.1244938682 |
- |
- |
kinase, putative [Ricinus communis] |