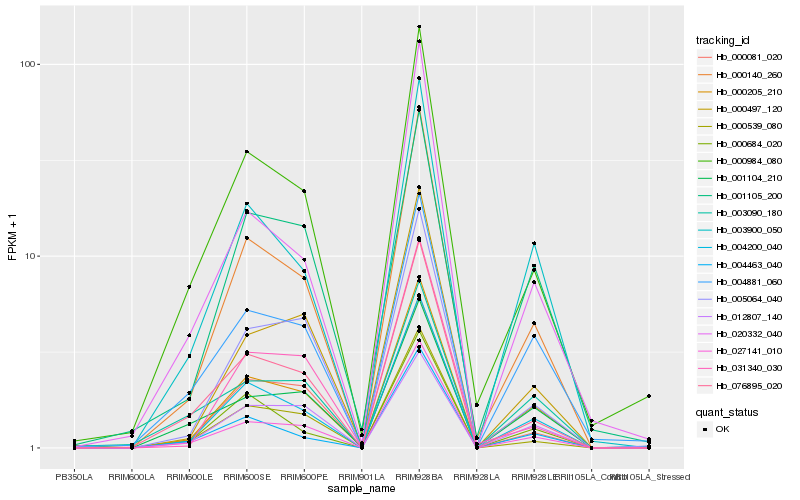
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000205_210 |
0.0 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein At4g27190-like isoform X1 [Citrus sinensis] |
| 2 |
Hb_004200_040 |
0.0578487635 |
- |
- |
laccase, putative [Ricinus communis] |
| 3 |
Hb_000140_260 |
0.0587219152 |
- |
- |
PREDICTED: uncharacterized protein LOC105642920 [Jatropha curcas] |
| 4 |
Hb_003900_050 |
0.0804615875 |
transcription factor |
TF Family: HSF |
Heat Stress Transcription Factor family protein [Populus trichocarpa] |
| 5 |
Hb_027141_010 |
0.0835859294 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 6 |
Hb_001105_200 |
0.0977047017 |
- |
- |
catalytic, putative [Ricinus communis] |
| 7 |
Hb_000984_080 |
0.0987499942 |
- |
- |
PREDICTED: receptor-like protein kinase HAIKU2 [Jatropha curcas] |
| 8 |
Hb_004881_060 |
0.0992436958 |
transcription factor |
TF Family: WRKY |
hypothetical protein POPTR_0005s22480g [Populus trichocarpa] |
| 9 |
Hb_000497_120 |
0.102281034 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_076895_020 |
0.1094120687 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g08850 isoform X2 [Jatropha curcas] |
| 11 |
Hb_000081_020 |
0.1099417506 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 1.1-like [Camelina sativa] |
| 12 |
Hb_001104_210 |
0.1124536768 |
- |
- |
hypothetical protein JCGZ_23620 [Jatropha curcas] |
| 13 |
Hb_005064_040 |
0.1128450208 |
- |
- |
hypothetical protein POPTR_0008s02680g [Populus trichocarpa] |
| 14 |
Hb_004463_040 |
0.1129793401 |
- |
- |
- |
| 15 |
Hb_000539_080 |
0.116020475 |
- |
- |
hypothetical protein POPTR_0039s00260g, partial [Populus trichocarpa] |
| 16 |
Hb_020332_040 |
0.1245982785 |
- |
- |
PREDICTED: alcohol dehydrogenase 1-like [Jatropha curcas] |
| 17 |
Hb_031340_030 |
0.125202444 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase IV.1 [Jatropha curcas] |
| 18 |
Hb_012807_140 |
0.125508788 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0009s01270g [Populus trichocarpa] |
| 19 |
Hb_003090_180 |
0.1272815752 |
- |
- |
PREDICTED: uncharacterized protein LOC105646674 [Jatropha curcas] |
| 20 |
Hb_000684_020 |
0.1276934632 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |