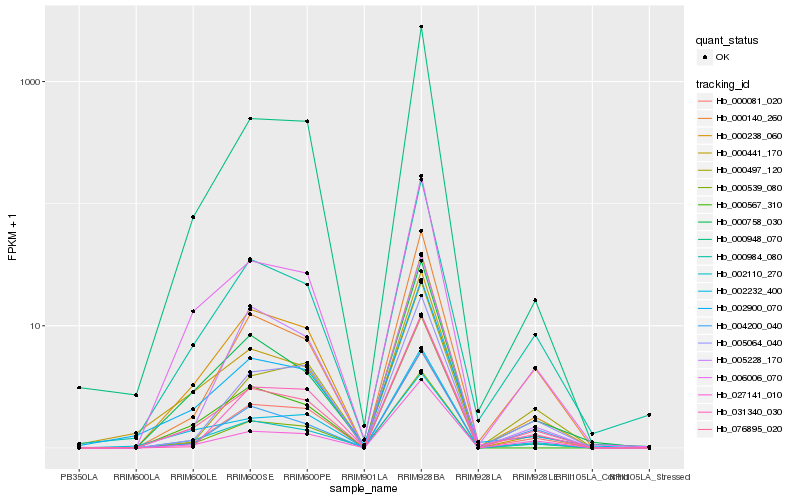
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000539_080 |
0.0 |
- |
- |
hypothetical protein POPTR_0039s00260g, partial [Populus trichocarpa] |
| 2 |
Hb_076895_020 |
0.0457023833 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g08850 isoform X2 [Jatropha curcas] |
| 3 |
Hb_000081_020 |
0.0594720927 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 1.1-like [Camelina sativa] |
| 4 |
Hb_000948_070 |
0.0696034956 |
- |
- |
hypothetical protein POPTR_0016s01720g [Populus trichocarpa] |
| 5 |
Hb_002110_270 |
0.0712817344 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH96 [Jatropha curcas] |
| 6 |
Hb_006006_070 |
0.0783400951 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 7 |
Hb_027141_010 |
0.0815241519 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 8 |
Hb_000140_260 |
0.083807186 |
- |
- |
PREDICTED: uncharacterized protein LOC105642920 [Jatropha curcas] |
| 9 |
Hb_005064_040 |
0.0843667838 |
- |
- |
hypothetical protein POPTR_0008s02680g [Populus trichocarpa] |
| 10 |
Hb_000984_080 |
0.0883410492 |
- |
- |
PREDICTED: receptor-like protein kinase HAIKU2 [Jatropha curcas] |
| 11 |
Hb_000758_030 |
0.0930277164 |
transcription factor |
TF Family: ERF |
Ethylene-responsive transcription factor, putative [Ricinus communis] |
| 12 |
Hb_031340_030 |
0.0952770898 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase IV.1 [Jatropha curcas] |
| 13 |
Hb_000238_060 |
0.1000473679 |
- |
- |
hypothetical protein JCGZ_24970 [Jatropha curcas] |
| 14 |
Hb_004200_040 |
0.1040620807 |
- |
- |
laccase, putative [Ricinus communis] |
| 15 |
Hb_002900_070 |
0.1051648986 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000497_120 |
0.1063507383 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_002232_400 |
0.107742731 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH25-like [Jatropha curcas] |
| 18 |
Hb_000441_170 |
0.1106632459 |
- |
- |
PREDICTED: uncharacterized protein LOC105636851 [Jatropha curcas] |
| 19 |
Hb_000567_310 |
0.1110164974 |
- |
- |
Miraculin precursor, putative [Ricinus communis] |
| 20 |
Hb_005228_170 |
0.1113142353 |
- |
- |
hypothetical protein JCGZ_24970 [Jatropha curcas] |