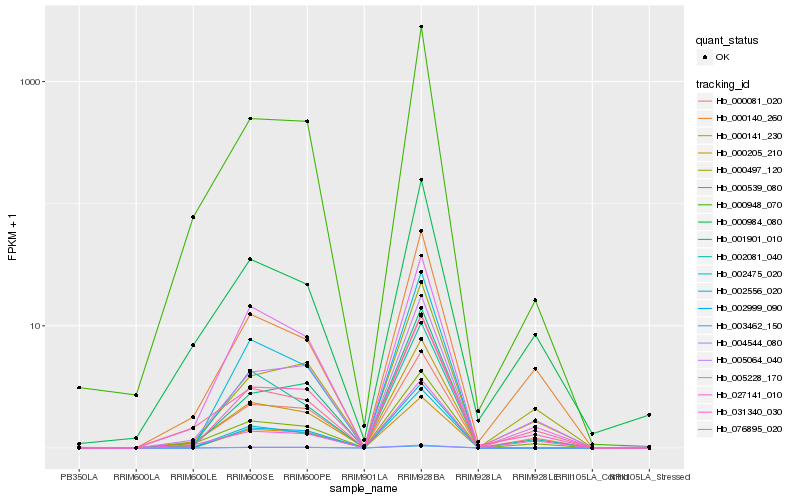
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_031340_030 |
0.0 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase IV.1 [Jatropha curcas] |
| 2 |
Hb_005064_040 |
0.0778694036 |
- |
- |
hypothetical protein POPTR_0008s02680g [Populus trichocarpa] |
| 3 |
Hb_000497_120 |
0.0829766996 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000081_020 |
0.0902729532 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 1.1-like [Camelina sativa] |
| 5 |
Hb_000539_080 |
0.0952770898 |
- |
- |
hypothetical protein POPTR_0039s00260g, partial [Populus trichocarpa] |
| 6 |
Hb_000140_260 |
0.1018183609 |
- |
- |
PREDICTED: uncharacterized protein LOC105642920 [Jatropha curcas] |
| 7 |
Hb_002999_090 |
0.1099085717 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000948_070 |
0.1107727664 |
- |
- |
hypothetical protein POPTR_0016s01720g [Populus trichocarpa] |
| 9 |
Hb_001901_010 |
0.1149031594 |
- |
- |
PREDICTED: alpha carbonic anhydrase 7-like [Jatropha curcas] |
| 10 |
Hb_003462_150 |
0.1159739826 |
- |
- |
PREDICTED: chitinase 1-like [Eucalyptus grandis] |
| 11 |
Hb_004544_080 |
0.116942933 |
- |
- |
lectin protein kinase [Populus trichocarpa] |
| 12 |
Hb_002475_020 |
0.1199927913 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At3g47570 isoform X1 [Populus euphratica] |
| 13 |
Hb_000141_230 |
0.1200325484 |
- |
- |
PREDICTED: 14 kDa proline-rich protein DC2.15-like [Camelina sativa] |
| 14 |
Hb_027141_010 |
0.1227707505 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 15 |
Hb_002556_020 |
0.1244835791 |
- |
- |
hypothetical protein JCGZ_12588 [Jatropha curcas] |
| 16 |
Hb_002081_040 |
0.1251698034 |
- |
- |
PREDICTED: nucleobase-ascorbate transporter 4 [Jatropha curcas] |
| 17 |
Hb_000205_210 |
0.125202444 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein At4g27190-like isoform X1 [Citrus sinensis] |
| 18 |
Hb_076895_020 |
0.1258625599 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g08850 isoform X2 [Jatropha curcas] |
| 19 |
Hb_005228_170 |
0.1275993702 |
- |
- |
hypothetical protein JCGZ_24970 [Jatropha curcas] |
| 20 |
Hb_000984_080 |
0.1303395871 |
- |
- |
PREDICTED: receptor-like protein kinase HAIKU2 [Jatropha curcas] |