| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
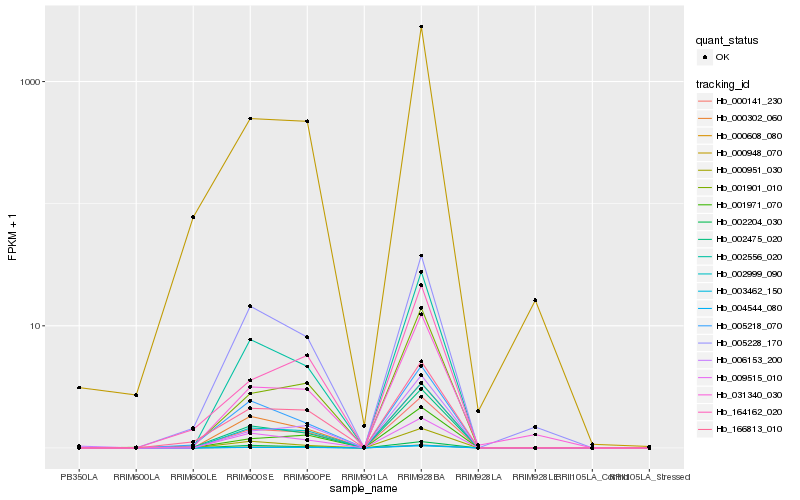
Hb_003462_150 |
0.0 |
- |
- |
PREDICTED: chitinase 1-like [Eucalyptus grandis] |
| 2 |
Hb_004544_080 |
0.0045116984 |
- |
- |
lectin protein kinase [Populus trichocarpa] |
| 3 |
Hb_000141_230 |
0.0192217823 |
- |
- |
PREDICTED: 14 kDa proline-rich protein DC2.15-like [Camelina sativa] |
| 4 |
Hb_000608_080 |
0.0421718998 |
- |
- |
hypothetical protein JCGZ_22216 [Jatropha curcas] |
| 5 |
Hb_002999_090 |
0.0499709932 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_002556_020 |
0.0690179099 |
- |
- |
hypothetical protein JCGZ_12588 [Jatropha curcas] |
| 7 |
Hb_002475_020 |
0.0724567473 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At3g47570 isoform X1 [Populus euphratica] |
| 8 |
Hb_000302_060 |
0.0752419266 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 9 |
Hb_001901_010 |
0.0818616523 |
- |
- |
PREDICTED: alpha carbonic anhydrase 7-like [Jatropha curcas] |
| 10 |
Hb_001971_070 |
0.0948151878 |
- |
- |
unnamed protein product [Coffea canephora] |
| 11 |
Hb_002204_030 |
0.1020142341 |
- |
- |
PREDICTED: uncharacterized protein LOC105764710 isoform X3 [Gossypium raimondii] |
| 12 |
Hb_166813_010 |
0.1020439742 |
- |
- |
hypothetical protein JCGZ_24970 [Jatropha curcas] |
| 13 |
Hb_009515_010 |
0.103061479 |
transcription factor |
TF Family: HB |
hypothetical protein JCGZ_15302 [Jatropha curcas] |
| 14 |
Hb_006153_200 |
0.1075108891 |
- |
- |
Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein [Theobroma cacao] |
| 15 |
Hb_000951_030 |
0.107760803 |
- |
- |
kinase, putative [Ricinus communis] |
| 16 |
Hb_031340_030 |
0.1159739826 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase IV.1 [Jatropha curcas] |
| 17 |
Hb_164162_020 |
0.121909831 |
- |
- |
PREDICTED: uncharacterized protein LOC105646964 [Jatropha curcas] |
| 18 |
Hb_005228_170 |
0.1219659203 |
- |
- |
hypothetical protein JCGZ_24970 [Jatropha curcas] |
| 19 |
Hb_000948_070 |
0.1236308143 |
- |
- |
hypothetical protein POPTR_0016s01720g [Populus trichocarpa] |
| 20 |
Hb_005218_070 |
0.1267302305 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor RAX3 isoform X2 [Populus euphratica] |