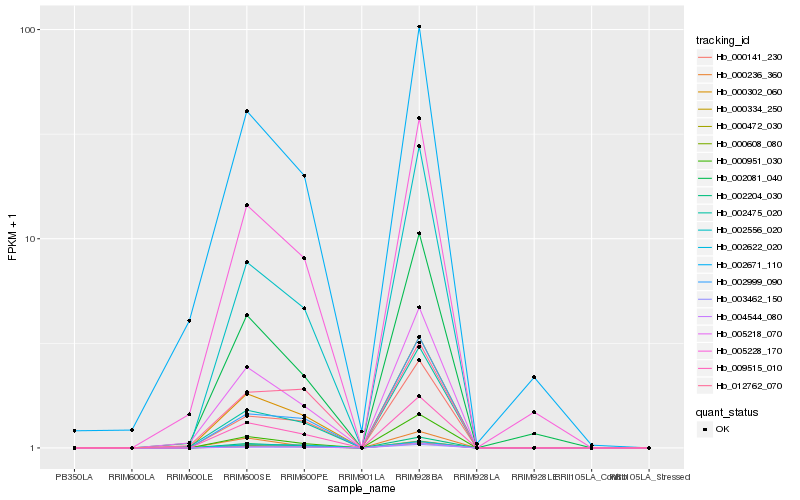
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009515_010 |
0.0 |
transcription factor |
TF Family: HB |
hypothetical protein JCGZ_15302 [Jatropha curcas] |
| 2 |
Hb_002204_030 |
0.0266964846 |
- |
- |
PREDICTED: uncharacterized protein LOC105764710 isoform X3 [Gossypium raimondii] |
| 3 |
Hb_000334_250 |
0.034971651 |
- |
- |
Polygalacturonase precursor, putative [Ricinus communis] |
| 4 |
Hb_000302_060 |
0.0411324941 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 5 |
Hb_005218_070 |
0.0798096951 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor RAX3 isoform X2 [Populus euphratica] |
| 6 |
Hb_000608_080 |
0.0824578883 |
- |
- |
hypothetical protein JCGZ_22216 [Jatropha curcas] |
| 7 |
Hb_000141_230 |
0.0846079745 |
- |
- |
PREDICTED: 14 kDa proline-rich protein DC2.15-like [Camelina sativa] |
| 8 |
Hb_005228_170 |
0.093317198 |
- |
- |
hypothetical protein JCGZ_24970 [Jatropha curcas] |
| 9 |
Hb_000951_030 |
0.0951761856 |
- |
- |
kinase, putative [Ricinus communis] |
| 10 |
Hb_002556_020 |
0.0976098511 |
- |
- |
hypothetical protein JCGZ_12588 [Jatropha curcas] |
| 11 |
Hb_000472_030 |
0.0999814087 |
- |
- |
- |
| 12 |
Hb_003462_150 |
0.103061479 |
- |
- |
PREDICTED: chitinase 1-like [Eucalyptus grandis] |
| 13 |
Hb_002475_020 |
0.1036946632 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At3g47570 isoform X1 [Populus euphratica] |
| 14 |
Hb_004544_080 |
0.1045533044 |
- |
- |
lectin protein kinase [Populus trichocarpa] |
| 15 |
Hb_002081_040 |
0.1131754199 |
- |
- |
PREDICTED: nucleobase-ascorbate transporter 4 [Jatropha curcas] |
| 16 |
Hb_002622_020 |
0.1140355868 |
- |
- |
- |
| 17 |
Hb_002671_110 |
0.1242303649 |
- |
- |
PREDICTED: remorin-like [Jatropha curcas] |
| 18 |
Hb_012762_070 |
0.1260588236 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein HOX3 [Jatropha curcas] |
| 19 |
Hb_000236_360 |
0.1263496003 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase FLS2 [Prunus mume] |
| 20 |
Hb_002999_090 |
0.1303547421 |
- |
- |
conserved hypothetical protein [Ricinus communis] |