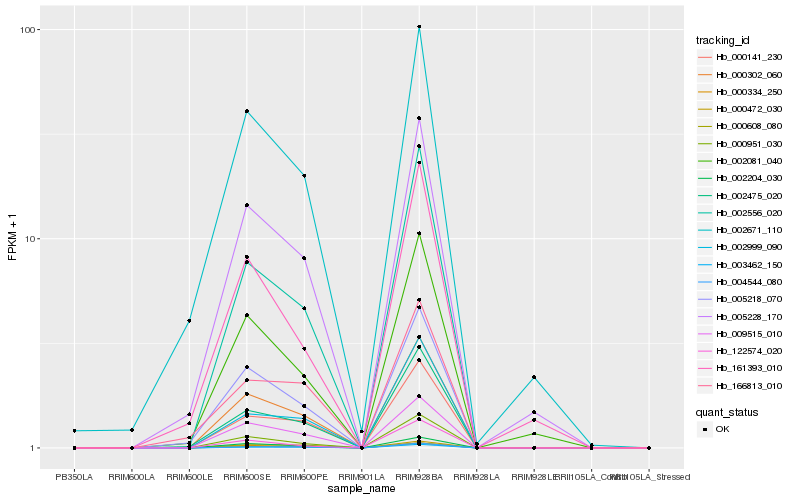
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000302_060 |
0.0 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 2 |
Hb_002204_030 |
0.0278779727 |
- |
- |
PREDICTED: uncharacterized protein LOC105764710 isoform X3 [Gossypium raimondii] |
| 3 |
Hb_009515_010 |
0.0411324941 |
transcription factor |
TF Family: HB |
hypothetical protein JCGZ_15302 [Jatropha curcas] |
| 4 |
Hb_000141_230 |
0.0562301978 |
- |
- |
PREDICTED: 14 kDa proline-rich protein DC2.15-like [Camelina sativa] |
| 5 |
Hb_002556_020 |
0.0566246221 |
- |
- |
hypothetical protein JCGZ_12588 [Jatropha curcas] |
| 6 |
Hb_000951_030 |
0.0619483638 |
- |
- |
kinase, putative [Ricinus communis] |
| 7 |
Hb_002475_020 |
0.0712387423 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At3g47570 isoform X1 [Populus euphratica] |
| 8 |
Hb_000608_080 |
0.0721608482 |
- |
- |
hypothetical protein JCGZ_22216 [Jatropha curcas] |
| 9 |
Hb_000334_250 |
0.0749345015 |
- |
- |
Polygalacturonase precursor, putative [Ricinus communis] |
| 10 |
Hb_003462_150 |
0.0752419266 |
- |
- |
PREDICTED: chitinase 1-like [Eucalyptus grandis] |
| 11 |
Hb_005218_070 |
0.0763667412 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor RAX3 isoform X2 [Populus euphratica] |
| 12 |
Hb_004544_080 |
0.0780896714 |
- |
- |
lectin protein kinase [Populus trichocarpa] |
| 13 |
Hb_005228_170 |
0.0911959897 |
- |
- |
hypothetical protein JCGZ_24970 [Jatropha curcas] |
| 14 |
Hb_002999_090 |
0.0921922814 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_002081_040 |
0.0994290249 |
- |
- |
PREDICTED: nucleobase-ascorbate transporter 4 [Jatropha curcas] |
| 16 |
Hb_166813_010 |
0.1204590182 |
- |
- |
hypothetical protein JCGZ_24970 [Jatropha curcas] |
| 17 |
Hb_002671_110 |
0.1247593221 |
- |
- |
PREDICTED: remorin-like [Jatropha curcas] |
| 18 |
Hb_000472_030 |
0.125697814 |
- |
- |
- |
| 19 |
Hb_122574_020 |
0.1288597536 |
- |
- |
hypothetical protein JCGZ_24076 [Jatropha curcas] |
| 20 |
Hb_161393_010 |
0.1305574723 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |