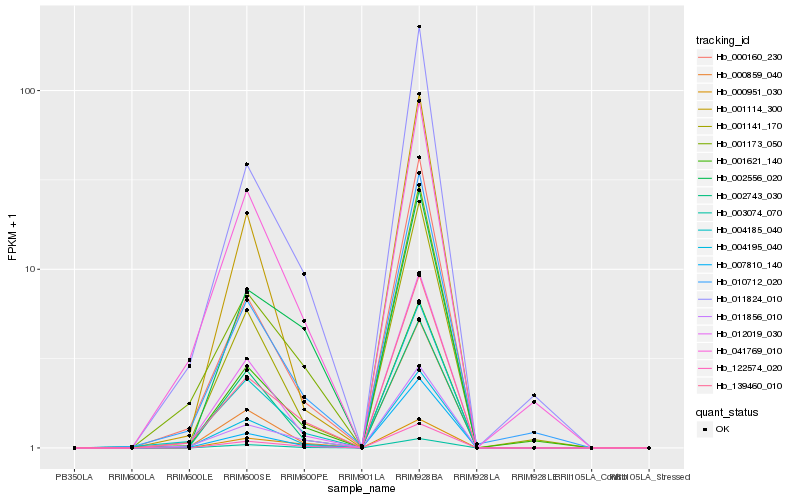
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_122574_020 |
0.0 |
- |
- |
hypothetical protein JCGZ_24076 [Jatropha curcas] |
| 2 |
Hb_003074_070 |
0.0666695017 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 3 |
Hb_000951_030 |
0.0681469045 |
- |
- |
kinase, putative [Ricinus communis] |
| 4 |
Hb_139460_010 |
0.073872913 |
- |
- |
PREDICTED: beta-glucosidase 18-like [Jatropha curcas] |
| 5 |
Hb_004195_040 |
0.0821065384 |
- |
- |
salicylic acid methyl transferase [Capsicum annuum] |
| 6 |
Hb_011856_010 |
0.0837152862 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 7 |
Hb_002556_020 |
0.090876229 |
- |
- |
hypothetical protein JCGZ_12588 [Jatropha curcas] |
| 8 |
Hb_012019_030 |
0.0909506505 |
- |
- |
hypothetical protein POPTR_0005s23360g [Populus trichocarpa] |
| 9 |
Hb_004185_040 |
0.0922928202 |
- |
- |
polygalacturonase, putative [Ricinus communis] |
| 10 |
Hb_011824_010 |
0.0983212282 |
- |
- |
Metalloendoproteinase 1 precursor, putative [Ricinus communis] |
| 11 |
Hb_001141_170 |
0.1013815276 |
- |
- |
PREDICTED: non-specific lipid-transfer protein-like protein At2g13820 [Jatropha curcas] |
| 12 |
Hb_001173_050 |
0.1034020525 |
- |
- |
DUF26 domain-containing protein 2 precursor, putative [Ricinus communis] |
| 13 |
Hb_001621_140 |
0.1075680871 |
- |
- |
PREDICTED: glycerol-3-phosphate acyltransferase 5 [Jatropha curcas] |
| 14 |
Hb_010712_020 |
0.1124320093 |
- |
- |
PREDICTED: caffeic acid 3-O-methyltransferase-like [Jatropha curcas] |
| 15 |
Hb_007810_140 |
0.1163659731 |
- |
- |
Ankyrin repeat-containing protein [Medicago truncatula] |
| 16 |
Hb_000859_040 |
0.116622594 |
- |
- |
Early nodulin 16 precursor, putative [Ricinus communis] |
| 17 |
Hb_002743_030 |
0.1172014833 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor RAX2 [Populus euphratica] |
| 18 |
Hb_000160_230 |
0.117717611 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB3-like [Jatropha curcas] |
| 19 |
Hb_001114_300 |
0.1182688236 |
- |
- |
hypothetical protein JCGZ_24970 [Jatropha curcas] |
| 20 |
Hb_041769_010 |
0.1190396751 |
- |
- |
hypothetical protein JCGZ_24970 [Jatropha curcas] |