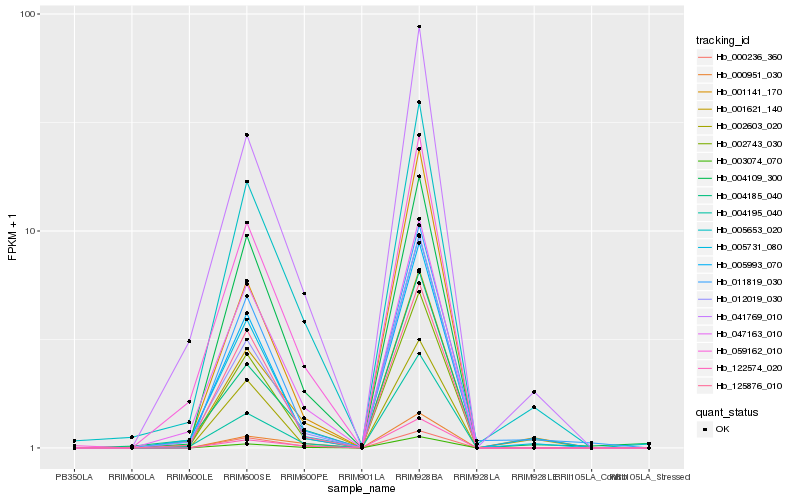
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003074_070 |
0.0 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 2 |
Hb_002743_030 |
0.0598313678 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor RAX2 [Populus euphratica] |
| 3 |
Hb_122574_020 |
0.0666695017 |
- |
- |
hypothetical protein JCGZ_24076 [Jatropha curcas] |
| 4 |
Hb_004195_040 |
0.0816209414 |
- |
- |
salicylic acid methyl transferase [Capsicum annuum] |
| 5 |
Hb_012019_030 |
0.0880590993 |
- |
- |
hypothetical protein POPTR_0005s23360g [Populus trichocarpa] |
| 6 |
Hb_004109_300 |
0.0905435029 |
- |
- |
hypothetical protein POPTR_0003s21800g [Populus trichocarpa] |
| 7 |
Hb_000951_030 |
0.0921648519 |
- |
- |
kinase, putative [Ricinus communis] |
| 8 |
Hb_047163_010 |
0.0933728326 |
- |
- |
PREDICTED: periaxin-like [Populus euphratica] |
| 9 |
Hb_059162_010 |
0.0945004619 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 10 |
Hb_001621_140 |
0.0953393745 |
- |
- |
PREDICTED: glycerol-3-phosphate acyltransferase 5 [Jatropha curcas] |
| 11 |
Hb_005731_080 |
0.0981497753 |
- |
- |
Anthranilate N-benzoyltransferase protein, putative [Ricinus communis] |
| 12 |
Hb_004185_040 |
0.0984333782 |
- |
- |
polygalacturonase, putative [Ricinus communis] |
| 13 |
Hb_002603_020 |
0.0998993038 |
- |
- |
PREDICTED: O-acetyl-ADP-ribose deacetylase MACROD2-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_125876_010 |
0.1002512996 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 15 |
Hb_011819_030 |
0.1072467318 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 16 |
Hb_000236_360 |
0.1115070739 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase FLS2 [Prunus mume] |
| 17 |
Hb_001141_170 |
0.1122612677 |
- |
- |
PREDICTED: non-specific lipid-transfer protein-like protein At2g13820 [Jatropha curcas] |
| 18 |
Hb_041769_010 |
0.112883579 |
- |
- |
hypothetical protein JCGZ_24970 [Jatropha curcas] |
| 19 |
Hb_005653_020 |
0.1140615962 |
transcription factor |
TF Family: C2H2 |
TRANSPARENT TESTA 1 protein, putative [Ricinus communis] |
| 20 |
Hb_005993_070 |
0.1141413878 |
- |
- |
lipid binding protein, putative [Ricinus communis] |