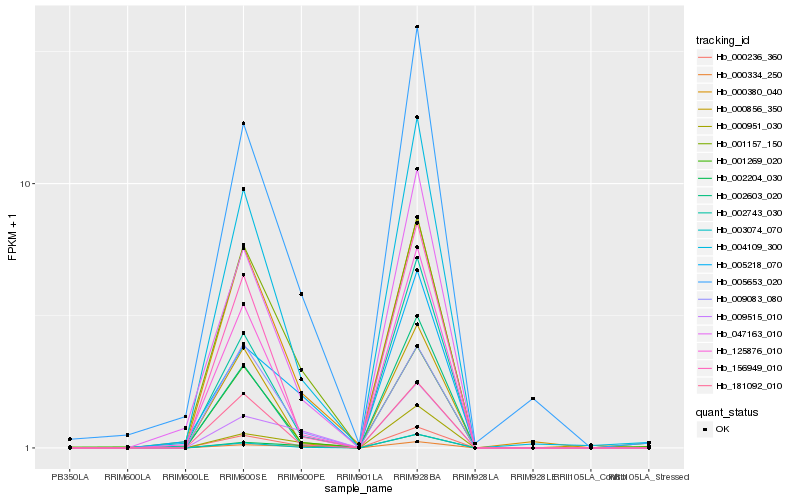
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000236_360 |
0.0 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase FLS2 [Prunus mume] |
| 2 |
Hb_004109_300 |
0.0767961088 |
- |
- |
hypothetical protein POPTR_0003s21800g [Populus trichocarpa] |
| 3 |
Hb_000380_040 |
0.0825718675 |
- |
- |
bel1 homeotic protein, putative [Ricinus communis] |
| 4 |
Hb_001157_150 |
0.0912747153 |
- |
- |
PREDICTED: transmembrane protein 45A-like [Jatropha curcas] |
| 5 |
Hb_047163_010 |
0.1008322738 |
- |
- |
PREDICTED: periaxin-like [Populus euphratica] |
| 6 |
Hb_125876_010 |
0.1008589697 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 7 |
Hb_181092_010 |
0.1030061538 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 8 |
Hb_156949_010 |
0.11116227 |
- |
- |
PREDICTED: terpene synthase 10-like [Jatropha curcas] |
| 9 |
Hb_003074_070 |
0.1115070739 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 10 |
Hb_002204_030 |
0.1133851477 |
- |
- |
PREDICTED: uncharacterized protein LOC105764710 isoform X3 [Gossypium raimondii] |
| 11 |
Hb_000334_250 |
0.1148836389 |
- |
- |
Polygalacturonase precursor, putative [Ricinus communis] |
| 12 |
Hb_002743_030 |
0.1170865046 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor RAX2 [Populus euphratica] |
| 13 |
Hb_009083_080 |
0.1193063804 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_005653_020 |
0.1194974671 |
transcription factor |
TF Family: C2H2 |
TRANSPARENT TESTA 1 protein, putative [Ricinus communis] |
| 15 |
Hb_002603_020 |
0.1260470999 |
- |
- |
PREDICTED: O-acetyl-ADP-ribose deacetylase MACROD2-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_001269_020 |
0.1261726908 |
- |
- |
PREDICTED: beta-Amyrin Synthase 2-like [Vitis vinifera] |
| 17 |
Hb_009515_010 |
0.1263496003 |
transcription factor |
TF Family: HB |
hypothetical protein JCGZ_15302 [Jatropha curcas] |
| 18 |
Hb_005218_070 |
0.1274711052 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor RAX3 isoform X2 [Populus euphratica] |
| 19 |
Hb_000951_030 |
0.1282098198 |
- |
- |
kinase, putative [Ricinus communis] |
| 20 |
Hb_000856_350 |
0.1300841895 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor 36 [Jatropha curcas] |